4ZN9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in complex with Oxabicyclic Heptene Sulfonate (OBHS) | | Descriptor: | Estrogen receptor, Nuclear receptor-interacting peptide, cyclohexa-2,5-dien-1-yl (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonate | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-05-04 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.215 Å) | | Cite: | Development of selective estrogen receptor modulator (SERM)-like activity through an indirect mechanism of estrogen receptor antagonism: defining the binding mode of 7-oxabicyclo[2.2.1]hept-5-ene scaffold core ligands.
Chemmedchem, 7, 2012
|
|
2R3W
 
 | | I84V HIV-1 protease in complex with a amino decorated pyrrolidine-based inhibitor | | Descriptor: | CHLORIDE ION, N,N'-(3S,4S)-PYRROLIDINE-3,4-DIYLBIS(4-AMINO-N-BENZYLBENZENESULFONAMIDE), Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-30 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
5GV0
 
 | | Crystal structure of the membrane-proximal domain of mouse lysosome-associated membrane protein 1 (LAMP-1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome-associated membrane glycoprotein 1, SULFATE ION | | Authors: | Tomabechi, Y, Ehara, H, Kukimoto-Niino, M, Shirouzu, M. | | Deposit date: | 2016-09-01 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lysosome-associated membrane proteins-1 and -2 (LAMP-1 and LAMP-2) assemble via distinct modes
Biochem.Biophys.Res.Commun., 479, 2016
|
|
5T9V
 
 | | Structure of rabbit RyR1 (Caffeine/ATP/Ca2+ dataset, class 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Clarke, O.B, des Georges, A, Zalk, R, Marks, A.R, Hendrickson, W.A, Frank, J. | | Deposit date: | 2016-09-09 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Basis for Gating and Activation of RyR1.
Cell, 167, 2016
|
|
9ELG
 
 | |
9ELQ
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11.1+S31 deletion spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
9ELO
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11+S31 deletion spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
6KMH
 
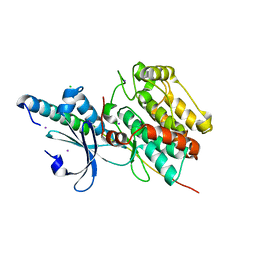 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
9ELE
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron KP.3.1.1 RBD in complex with human ACE2 (local refinement of RBD and hACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
9ELN
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11+S31 deletion spike protein (one RBD up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-05-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
3DN2
 
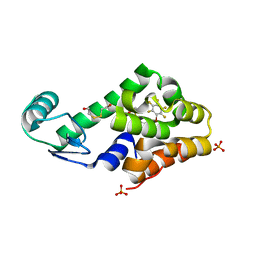 | | Bromopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1-bromo-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
5TL8
 
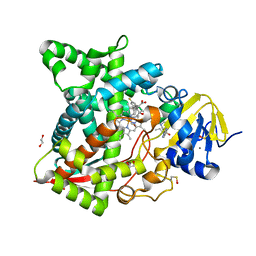 | | Naegleria fowleri CYP51-posaconazole complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, POSACONAZOLE, ... | | Authors: | Podust, L.M, Jennings, G, Calvet-Alvarez, C, Debnath, A. | | Deposit date: | 2016-10-10 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of the Naegleria fowleri CYP51 at 1.7 Angstroms resolution
To be published
|
|
9ELM
 
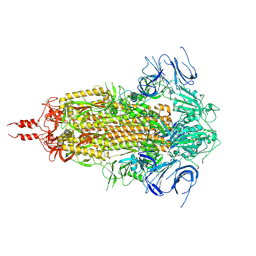 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11 spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
2R38
 
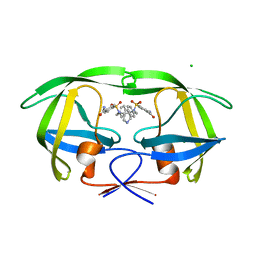 | | I84V HIV-1 protease mutant in complex with a carbamoyl decorated pyrrolidine-based inhibitor | | Descriptor: | 4,4'-{(3S,4S)-PYRROLIDINE-3,4-DIYLBIS[(BENZYLIMINO)SULFONYL]}DIBENZAMIDE, CHLORIDE ION, Protease | | Authors: | Boettcher, J, Blum, A, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-08-29 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Kinetic Analysis of Pyrrolidine-Based Inhibitors of the Drug-Resistant Ile84Val Mutant of HIV-1 Protease
J.Mol.Biol., 383, 2008
|
|
7WRS
 
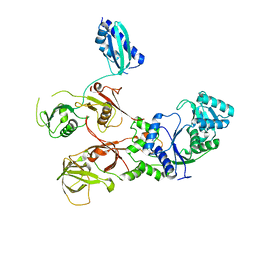 | |
9ELJ
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11+Q493E+S31deletion spike protein (one RBD up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
4PY3
 
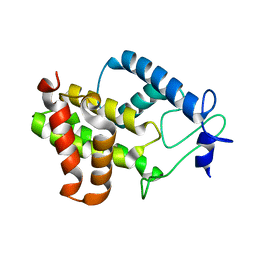 | |
9ELL
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11 spike protein (one RBD up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
9ELK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1.11+Q493E+S31deletion spike protein (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Feng, Z, Huang, J, Ward, A.B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-06-04 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Structural and functional insights into the evolution of SARS-CoV-2 KP.3.1.1 spike protein.
Cell Rep, 44, 2025
|
|
4CTC
 
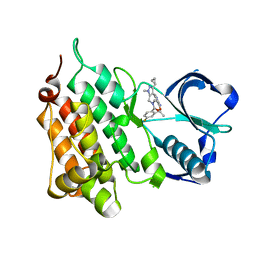 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with the inhibitor 7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17- dihydro-1H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecin-15(10H)-one | | Descriptor: | (10R)-7-amino-3-cyclopropyl-12-fluoro-1,10,16-trimethyl-16,17-dihydro-1H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecin-15(10H)-one, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
3DN8
 
 | | Iodopentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant (seleno version) | | Descriptor: | 1,2,3,4,5-pentafluoro-6-iodobenzene, 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
1WH4
 
 | | Solution structure of the DEATH domain of Interleukin-1 receptor-associated kinase4 (IRAK4) from Mus musculus | | Descriptor: | interleukin-1 receptor-associated kinase 4 | | Authors: | Nameki, N, Tomizawa, T, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DEATH domain of Interleukin-1 receptor-associated kinase4 (IRAK4) from Mus musculus
To be Published
|
|
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
9J6G
 
 | | Cryo-EM structure of Bat SARS-like coronavirus Khosta-1 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pan, X.Q, Li, L.J, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-08-15 | | Release date: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of receptor recognition by Khosta-1/Khosta-2 spike proteins
To Be Published
|
|
7EC9
 
 | |
