2V0R
 
 | | crystal structure of a hairpin exchange variant (LTx) of the targeting LINE-1 retrotransposon endonuclease | | Descriptor: | LTX, SULFATE ION | | Authors: | Repanas, K, Zingler, N, Layer, L.E, Schumann, G.G, Perrakis, A, Weichenrieder, O. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determinants for DNA Target Structure Selectivity of the Human Line-1 Retrotransposon Endonuclease
Nucleic Acids Res., 35, 2007
|
|
6GT7
 
 | |
4BEB
 
 | | MUTANT (K220E) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, TYPE I RESTRICTION ENZYME HSDR | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
1D4B
 
 | | CIDE-N DOMAIN OF HUMAN CIDE-B | | Descriptor: | HUMAN CELL DEATH-INDUCING EFFECTOR B | | Authors: | Lugovskoy, A, Zhou, P, Chou, J, McCarty, J, Li, P, Wagner, G. | | Deposit date: | 1999-10-02 | | Release date: | 1999-12-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CIDE-N domain of CIDE-B and a model for CIDE-N/CIDE-N interactions in the DNA fragmentation pathway of apoptosis.
Cell(Cambridge,Mass.), 99, 1999
|
|
6IBC
 
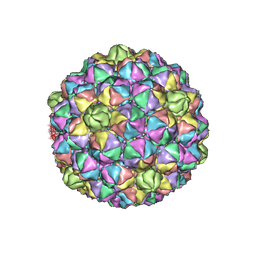 | | Thermophage P23-45 procapsid | | Descriptor: | Major head protein | | Authors: | Bayfield, O.W, Klimuk, E, Winkler, D.C, Hesketh, E.L, Chechik, M, Cheng, N, Dykeman, E.C, Minakhin, L, Ranson, N.A, Severinov, K, Steven, A.C, Antson, A.A. | | Deposit date: | 2018-11-29 | | Release date: | 2019-02-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.39 Å) | | Cite: | Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5DMP
 
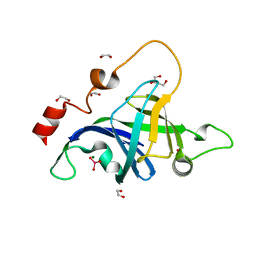 | | Structure of the Archaeal NHEJ Phosphoesterase from Methanocella paludicola. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
1JUA
 
 | |
1Z6A
 
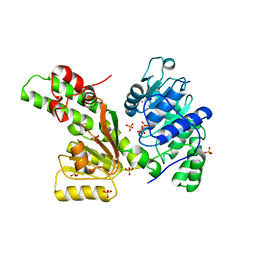 | | Sulfolobus solfataricus SWI2/SNF2 ATPase core domain | | Descriptor: | Helicase of the snf2/rad54 family, MERCURY (II) ION, PHOSPHATE ION | | Authors: | Duerr, H, Koerner, C, Mueller, M, Hickmann, V, Hopfner, K.P. | | Deposit date: | 2005-03-22 | | Release date: | 2005-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA
Cell(Cambridge,Mass.), 121, 2005
|
|
6ES8
 
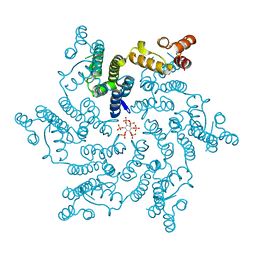 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
2LQ6
 
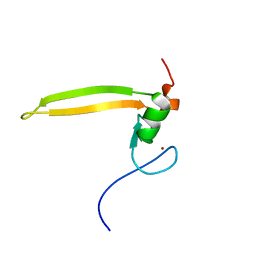 | | Solution structure of BRD1 PHD2 finger | | Descriptor: | Bromodomain-containing protein 1, ZINC ION | | Authors: | Liu, L, Wu, J. | | Deposit date: | 2012-02-25 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an atypical PHD finger in BRPF2 and its interaction with DNA
J.Struct.Biol., 180, 2012
|
|
4C8H
 
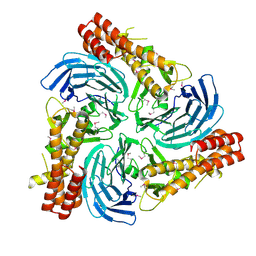 | |
5BQ5
 
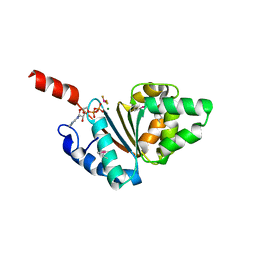 | | Crystal structure of the IstB AAA+ domain bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Insertion sequence IS5376 putative ATP-binding protein, ... | | Authors: | Arias-Palomo, E, Berger, J.M. | | Deposit date: | 2015-05-28 | | Release date: | 2015-09-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Atypical AAA+ ATPase Assembly Controls Efficient Transposition through DNA Remodeling and Transposase Recruitment.
Cell, 162, 2015
|
|
6EIH
 
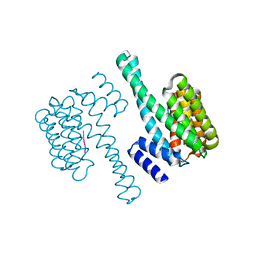 | |
2MX0
 
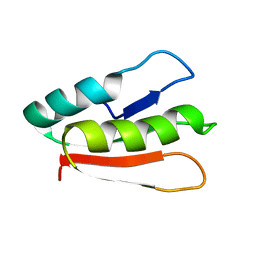 | | Solution structure of HP0268 from Helicobacter pylori | | Descriptor: | Uncharacterized protein HP_0268 | | Authors: | Lee, K.Y. | | Deposit date: | 2014-12-05 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-based functional identification of Helicobacter pylori HP0268 as a nuclease with both DNA nicking and RNase activities
Nucleic Acids Res., 43, 2015
|
|
6FLF
 
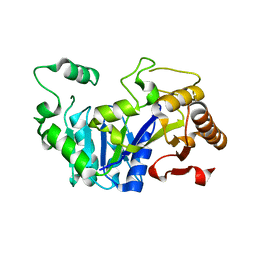 | |
6FM1
 
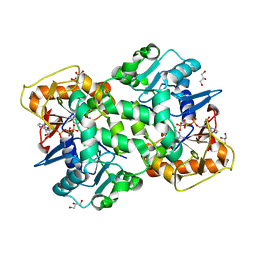 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATPanddGMP at 2.3 Angstrom resolution | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
1S16
 
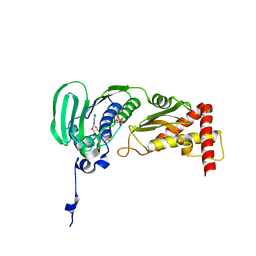 | | Crystal Structure of E. coli Topoisomerase IV ParE 43kDa subunit complexed with ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Wei, Y, Gross, C.H. | | Deposit date: | 2004-01-05 | | Release date: | 2004-05-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Escherichia coli topoisomerase IV ParE subunit (24 and 43 kilodaltons): a single residue dictates differences in novobiocin potency against topoisomerase IV and DNA gyrase.
Antimicrob.Agents Chemother., 48, 2004
|
|
6FM0
 
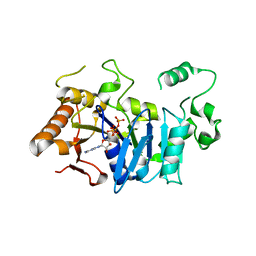 | | Deoxyguanylosuccinate synthase (DgsS) and ATP structure at 1.7 Angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2018-01-29 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
2N9E
 
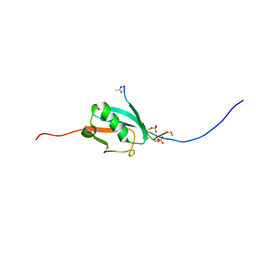 | |
2LLA
 
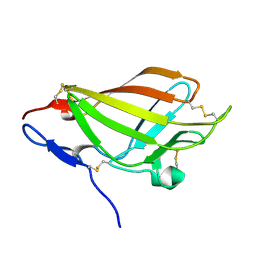 | | NMR solution structure ensemble of domain 11 of the echidna M6P/IGF2R receptor | | Descriptor: | Mannose-6-phosphate/insulin-like growth factor II receptor | | Authors: | Strickland, M, Crump, M.P, Williams, C, Rezgui, D, Ellis, R.Z, Hoppe, H, Frago, S, Prince, S.N, Zaccheo, O.J, Forbes, B.E, Jones, E, Hassan, A.Z, Wattana-Amorn, P. | | Deposit date: | 2011-11-05 | | Release date: | 2012-11-07 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | An exon splice enhancer primes IGF2:IGF2R binding site structure and function evolution.
Science, 338, 2012
|
|
1XVK
 
 | |
1XVR
 
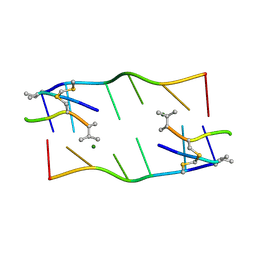 | | echinomycin (CGTACG)2 complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*CP*GP*TP*AP*CP*G)-3', ECHINOMYCIN, ... | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M. | | Deposit date: | 2004-10-28 | | Release date: | 2005-04-12 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of Complexes between Echinomycin and Duplex DNA.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2GK4
 
 | |
1XVN
 
 | | echinomycin (ACGTACGT)2 complex | | Descriptor: | 2-CARBOXYQUINOXALINE, 5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3', ECHINOMYCIN, ... | | Authors: | Cuesta-Seijo, J.A, Sheldrick, G.M. | | Deposit date: | 2004-10-28 | | Release date: | 2005-04-12 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of Complexes between Echinomycin and Duplex DNA.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3KKR
 
 | |
