2H7E
 
 | |
2KR2
 
 | |
2KSC
 
 | |
1CUW
 
 | | CUTINASE, G82A, A85F, V184I, A185L, L189F MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUE
 
 | | CUTINASE, Q121L MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUC
 
 | |
1CUX
 
 | | CUTINASE, L114Y MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUB
 
 | |
1CUG
 
 | | CUTINASE, R17E, N172K MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUD
 
 | |
1CUU
 
 | | CUTINASE, A199C MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUH
 
 | | CUTINASE, R196E MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUF
 
 | | CUTINASE, R156L MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
4MKI
 
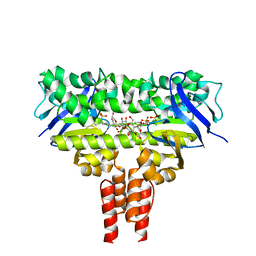 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
4BGV
 
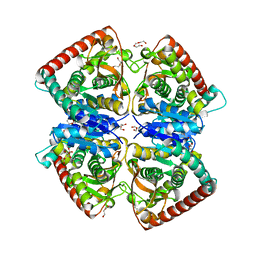 | |
4BGU
 
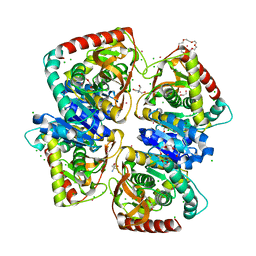 | | 1.50 A resolution structure of the malate dehydrogenase from Haloferax volcanii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Talon, R, Madern, D, Girard, E. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.487 Å) | | Cite: | Insight Into Structural Evolution of Extremophilic Proteins
To be Published
|
|
2JMV
 
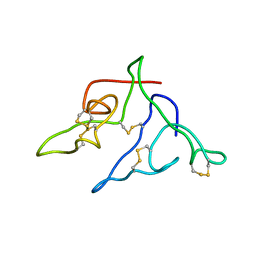 | |
2LBO
 
 | |
2GVS
 
 | | NMR solution structure of CSPsg4 | | Descriptor: | chemosensory protein CSP-sg4 | | Authors: | Tomaselli, S, Crescenzi, O, Sanfelice, D, Ab, E, Tancredi, T, Picone, D. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-12 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Chemosensory Protein from the Desert Locust Schistocerca gregaria(,).
Biochemistry, 45, 2006
|
|
1CUA
 
 | | CUTINASE, N172K MUTANT | | Descriptor: | CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
1CUI
 
 | | CUTINASE, S120A MUTANT | | Descriptor: | CUTINASE | | Authors: | Martinez, C, Cambillau, C. | | Deposit date: | 1995-11-16 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics of Fusarium solani cutinase investigated through structural comparison among different crystal forms of its variants.
Proteins, 26, 1996
|
|
2L23
 
 | |
2LR3
 
 | |
2L55
 
 | |
2LVY
 
 | |
