6S6S
 
 | |
5JRI
 
 | | Structure of an oxidoreductase SeMet-labelled from Synechocystis sp. PCC6803 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase, ... | | Authors: | Buey, R.M, de Pereda, J.M, Balsera, M. | | Deposit date: | 2016-05-06 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Unprecedented pathway of reducing equivalents in a diflavin-linked disulfide oxidoreductase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3PQB
 
 | | The crystal structure of pregilvocarcin in complex with GilR, an oxidoreductase that catalyzes the terminal step of gilvocarcin biosynthesis | | Descriptor: | (1R)-1,4-anhydro-6-deoxy-1-[(6R)-8-ethenyl-1,6-dihydroxy-10,12-dimethoxy-6H-dibenzo[c,h]chromen-4-yl]-D-galactitol, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase | | Authors: | Noinaj, N, Bosserman, M.A, Schickli, M.A, Kharel, M.K, Rohr, J, Buchanan, S.K. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | The Crystal Structure and Mechanism of an Unusual Oxidoreductase, GilR, Involved in Gilvocarcin V Biosynthesis.
J.Biol.Chem., 286, 2011
|
|
1NG3
 
 | | Complex of ThiO (glycine oxidase) with acetyl-glycine | | Descriptor: | ACETYLAMINO-ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Glycine oxidase, ... | | Authors: | Settembre, E.C, Dorrestein, P.C, Park, J, Augustine, A, Begley, T.P, Ealick, S.E. | | Deposit date: | 2002-12-16 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Studies on ThiO, a Glycine Oxidase Essential for Thiamin Biosynthesis in Bacillus subtilis
Biochemistry, 42, 2003
|
|
2I0K
 
 | |
2B76
 
 | | E. coli Quinol fumarate reductase FrdA E49Q mutation | | Descriptor: | CITRATE ANION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Maklashina, E, Iverson, T.M, Sher, Y, Kotlyar, V, Mirza, O, Andrell, J, Hudson, J.M, Armstrong, F.A, Cecchini, G. | | Deposit date: | 2005-10-03 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fumarate Reductase and Succinate Oxidase Activity of Escherichia coli Complex II Homologs Are Perturbed Differently by Mutation of the Flavin Binding Domain
J.Biol.Chem., 281, 2006
|
|
7ESC
 
 | | FmnB complexed with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7ESB
 
 | | FmnB complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Zheng, Y.H, Cheng, W. | | Deposit date: | 2021-05-09 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
7F2U
 
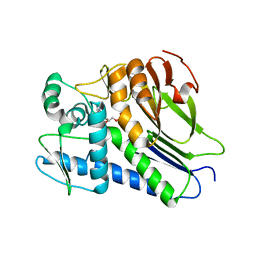 | | FmnB complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FAD:protein FMN transferase, MAGNESIUM ION | | Authors: | Cheng, W, Zheng, Y.H. | | Deposit date: | 2021-06-14 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Structural insights into the catalytic and inhibitory mechanisms of the flavin transferase FmnB in Listeria monocytogenes.
MedComm (2020), 3, 2022
|
|
1F6M
 
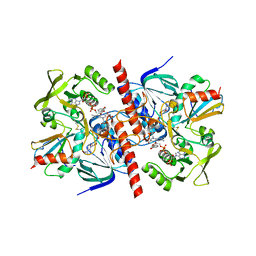 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
5URG
 
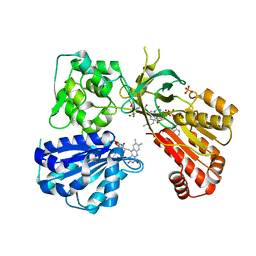 | | rat CYPOR D632F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URH
 
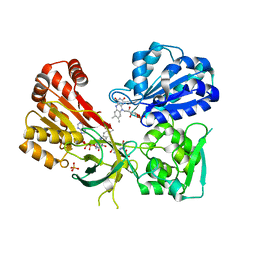 | | CYPOR/D632A with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
6CY8
 
 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-05 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.729 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
5URI
 
 | | Rat CYPOR/D632A with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URD
 
 | | wild type rat CYPOR bound with NADP+ - oxidized form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5URE
 
 | | Wild type rat CYPOR bound with NADP+ - reduced form | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Studies of Asp632 Mutants and Fully Reduced NADPH-Cytochrome P450 Oxidoreductase Define the Role of Asp632 Loop Dynamics in the Control of NADPH Binding and Hydride Transfer.
Biochemistry, 57, 2018
|
|
5IOR
 
 | | Flavin-dependent thymidylate synthase in complex with FAD and 2'-deoxyuridine-5'-monosulfate | | Descriptor: | 2'-deoxy-5'-O-sulfouridine, FLAVIN-ADENINE DINUCLEOTIDE, RIBOFLAVIN, ... | | Authors: | Bernard, S.M, Stull, F.W, Smith, J.L. | | Deposit date: | 2016-03-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Deprotonations in the Reaction of Flavin-Dependent Thymidylate Synthase.
Biochemistry, 55, 2016
|
|
6TE1
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol | | Descriptor: | 5-[4-cyclobutyl-1-[2-(4-piperidin-4-yloxyphenoxy)ethyl]imidazol-2-yl]-4-methyl-thieno[3,2-b]pyrrole, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Pasqualato, S, Cecatiello, V. | | Deposit date: | 2019-11-11 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of Reversible Inhibitors of KDM1A Efficacious in Acute Myeloid Leukemia Models.
Acs Med.Chem.Lett., 11, 2020
|
|
6CXT
 
 | | Crystal structure of FAD-dependent dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase fold protein, Butyryl-CoA dehydrogenase, ... | | Authors: | Agarwal, V. | | Deposit date: | 2018-04-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into Thiotemplated Pyrrole Biosynthesis Gained from the Crystal Structure of Flavin-Dependent Oxidase in Complex with Carrier Protein.
Biochemistry, 58, 2019
|
|
3GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, OXIDIZED TRYPANOTHIONE COMPLEX | | Descriptor: | 2-AMINO-4-[4-(4-AMINO-4-CARBOXY-BUTYRYLAMINO)-5,8,19,22-TETRAOXO-1,2-DITHIA-6,9,13,18,21-PENTAAZA-CYCLOTETRACOS-23-YLCARBAMOYL]-BUTYRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
4JAY
 
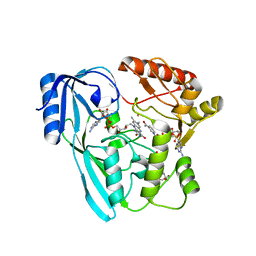 | | Crystal structure of P. aeruginosa MurB in complex with NADP+ | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chen, M.W, Lohkamp, B, Schnell, R, Lescar, J, Schneider, G. | | Deposit date: | 2013-02-19 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Substrate Channel Flexibility in Pseudomonas aeruginosa MurB Accommodates Two Distinct Substrates.
Plos One, 8, 2013
|
|
3S1C
 
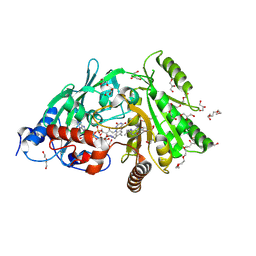 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1D
 
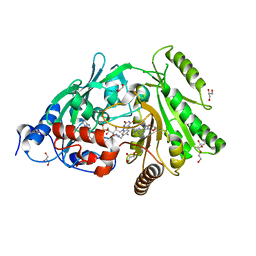 | | Glu381Ser mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1F
 
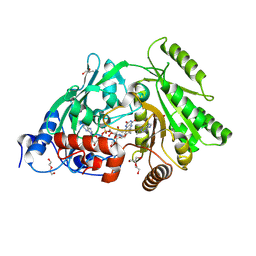 | | Asp169Glu mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3S1E
 
 | | Pro427Gln mutant of maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
