7O7E
 
 | | Crystal structure of rsEGFP2 mutant V151L in the fluorescent on-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7W
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state the determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
5UDL
 
 | | IFIT1 N216A monomeric mutant (L457E/L464E) with m7Gppp-AAAA (anti conformation of cap) | | Descriptor: | CALCIUM ION, Interferon-induced protein with tetratricopeptide repeats 1, RNA (5'-D(*(GTA))-R(P*AP*AP*A)-3'), ... | | Authors: | Abbas, Y.M, Martinez-Montero, S, Damha, M.J, Nagar, B. | | Deposit date: | 2016-12-27 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7O7V
 
 | | Crystal structure of rsEGFP2 mutant V151A in the fluorescent on-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7H
 
 | | Crystal structure of rsEGFP2 mutant V151L in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7X
 
 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by serial femtosecond crystallography at room temperature | | Descriptor: | Green fluorescent protein | | Authors: | Hadjidemetriou, K, Coquelle, N, Barends, T.R.M, Schlichting, I, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7O7C
 
 | | Crystal structure of rsEGFP2 mutant V151A in the non-fluorescent off-state determined by synchrotron radiation at 100K | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Woodhouse, J, Adam, V, Hadjidemetriou, K, Colletier, J.-P, Weik, M. | | Deposit date: | 2021-04-13 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Control of Off-State Heterogeneity in a Photoswitchable Fluorescent Protein Provides Switching Contrast Enhancement.
Chemphyschem, 23, 2022
|
|
7QOD
 
 | |
5YSE
 
 | | Crystal structure of beta-1,2-glucooligosaccharide binding protein in complex with sophorotetraose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lin1841 protein, MAGNESIUM ION, ... | | Authors: | Abe, K, Nakajima, M, Taguchi, H, Arakawa, T, Fushinobu, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic insights into beta-1,2-glucooligosaccharide capture by a solute-binding protein inListeria innocua.
J. Biol. Chem., 293, 2018
|
|
7OOH
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818 | | Descriptor: | 4-IODOPYRAZOLE, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818
To Be Published
|
|
5C0E
 
 | | HLA-A02 carrying YLGGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
7OOG
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023823 | | Descriptor: | 4-bromanylpyridin-2-amine, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023823
To Be Published
|
|
7OOE
 
 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with Z321318226 | | Descriptor: | AMP PHOSPHORAMIDATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-05-27 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of P. falciparum Hsp70-x nucleotide binding domain with small molecule ligands
To Be Published
|
|
7OP1
 
 | | Cryo-EM structure of P5B-ATPase E2PiAlF/SPM | | Descriptor: | Cation-transporting ATPase, MAGNESIUM ION, SPERMINE, ... | | Authors: | Li, P, Gourdon, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
7OTJ
 
 | | Crystal structure of Pif1 helicase from Candida albicans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(P*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rety, S, Xi, X.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural study of the function of Candida Albicans Pif1.
Biochem.Biophys.Res.Commun., 567, 2021
|
|
7OR9
 
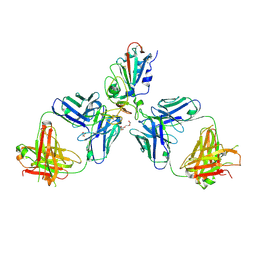 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and COVOX-278 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-06-04 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum.
Cell, 184, 2021
|
|
7P31
 
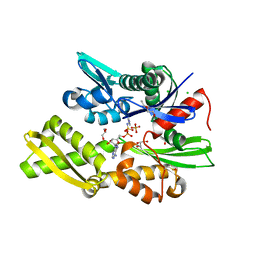 | | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with NCL-00023818 | | Descriptor: | 4-IODOPYRAZOLE, AMP PHOSPHORAMIDATE, CHLORIDE ION, ... | | Authors: | Mohamad, N, O'Donoghue, A, Kantsadi, A.L, Vakonakis, I. | | Deposit date: | 2021-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Plasmodium falciparum Hsp70-x chaperone nucleotide binding domain in complex with small molecule ligands
To Be Published
|
|
7QLJ
 
 | |
5SWD
 
 | | Structure of the adenine riboswitch aptamer domain in an intermediate-bound state | | Descriptor: | ADENINE, MAGNESIUM ION, Vibrio vulnificus strain 93U204 chromosome II, ... | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
5SWE
 
 | | Ligand-bound structure of adenine riboswitch aptamer domain converted in crystal from its ligand-free state using ligand mixing serial femtosecond crystallography | | Descriptor: | ADENINE, Vibrio vulnificus strain 93U204 chromosome II, adenine riboswitch aptamer domain | | Authors: | Stagno, J.R, Wang, Y.-X, Liu, Y, Bhandari, Y.R, Conrad, C.E, Nelson, G, Li, C, Wendel, D.R, White, T.A, Barty, A, Tuckey, R.A, Zatsepin, N.A, Grant, T.D, Fromme, P, Tan, K, Ji, X, Spence, J.C.H. | | Deposit date: | 2016-08-08 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography.
Nature, 541, 2017
|
|
7QI3
 
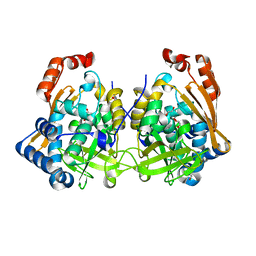 | | Structure of Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Arylamine N-acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lowe, E.D, Kotomina, E, Karagianni, E, Boukouvala, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase is structurally, functionally and phylogenetically distinct from its N-acetyltransferase (NAT) homologues.
Febs J., 290, 2023
|
|
7QJR
 
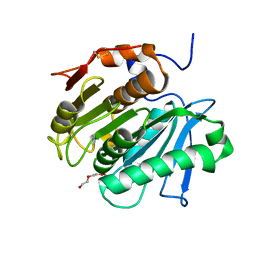 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
7QJT
 
 | | Crystal structure of a cutinase enzyme from Thermobifida cellulosilytica TB100 (711) | | Descriptor: | GLYCEROL, MAGNESIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJS
 
 | | Crystal structure of a cutinase enzyme from Thermobifida fusca YX (705) | | Descriptor: | Cutinase 2, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zahn, M, Shakespeare, T.J, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
