1NVL
 
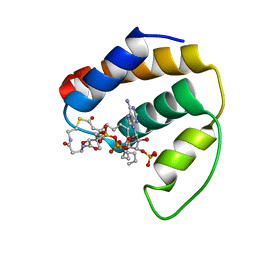 | | RDC-refined NMR structure of bovine Acyl-coenzyme A Binding Protein, ACBP, in complex with palmitoyl-coenzyme A | | Descriptor: | Acyl-CoA-binding protein, COENZYME A, PALMITIC ACID | | Authors: | Lerche, M.H, Kragelund, B.B, Redfield, C, Poulsen, F.M. | | Deposit date: | 2003-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RDC-refined NMR structure of bovine Acyl-coenzyme A Binding Protein, ACBP, in complex with palmitoyl-coenzyme A
To be Published, 2003
|
|
1QK7
 
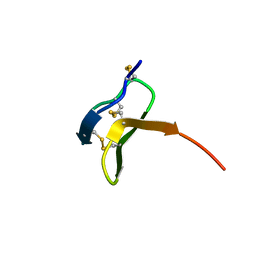 | |
1MUV
 
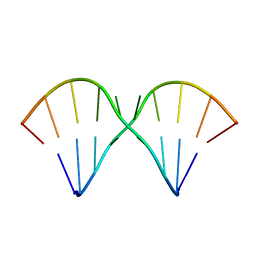 | | Sheared A(anti)-A(anti) Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of 5'(rGGCAAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Schroeder, S.J, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheared Aanti-Aanti Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of
5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1AYJ
 
 | |
7ZNF
 
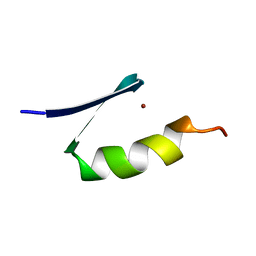 | |
8X4F
 
 | |
2RPI
 
 | |
1PCO
 
 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | HYDROXIDE ION, PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1PCN
 
 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | HYDROXIDE ION, PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
3MEF
 
 | |
2V93
 
 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
1OEF
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 263-286, NMR, 5 STRUCTURES AT PH 4.8, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1OEG
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 267-289, NMR, 5 STRUCTURES AT PH 6.0, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1HDP
 
 | | SOLUTION STRUCTURE OF A POU-SPECIFIC HOMEODOMAIN: 3D-NMR STUDIES OF HUMAN B-CELL TRANSCRIPTION FACTOR OCT-2 | | Descriptor: | OCT-2 POU HOMEODOMAIN | | Authors: | Sivaraja, M, Botfield, M.C, Mueller, M, Jancso, A, Weiss, M.A. | | Deposit date: | 1994-03-08 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a POU-specific homeodomain: 3D-NMR studies of human B-cell transcription factor Oct-2.
Biochemistry, 33, 1994
|
|
1Q7O
 
 | | Determination of f-MLF-OH Peptide Structure with solid-state magic-angle spinning NMR Spectroscopy | | Descriptor: | chemotactic peptide | | Authors: | Rienstra, C.M, Tucker-Kellogg, L, Jaroniec, C.P, Hohwy, M, Reif, B, McMahon, M.T, Tidor, B, Lozano-Perez, T, Griffin, R.G. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | De novo determination of peptide structure with solid-state magic-angle spinning NMR Spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1OLG
 
 | |
6A7Y
 
 | | Solution structure of an intermolecular leaped V-shape G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*A)-3') | | Authors: | Wan, C.J, Zhang, N. | | Deposit date: | 2018-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an asymmetric intermolecular leaped V-shape G-quadruplex: selective recognition of the d(G2NG3NG4) sequence motif by a short linear G-rich DNA probe.
Nucleic Acids Res., 47, 2019
|
|
1ODQ
 
 | |
1BMX
 
 | |
1GPS
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1GPT
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-H THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1ODP
 
 | |
1ODR
 
 | |
1OLH
 
 | |
6QBJ
 
 | |
