6RW8
 
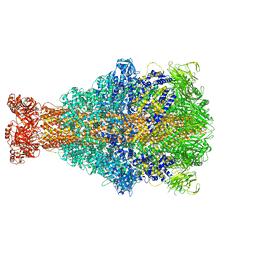 | | Cryo-EM structure of Xenorhabdus nematophila XptA1 | | Descriptor: | A component of insecticidal toxin complex (Tc) | | Authors: | Roderer, D, Leidreiter, F, Gatsogiannis, C, Meusch, D, Benz, R, Raunser, S. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Common architecture of Tc toxins from human and insect pathogenic bacteria.
Sci Adv, 5, 2019
|
|
4UWV
 
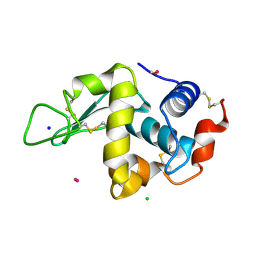 | | Lysozyme soaked with a ruthenium based CORM with a pyridine ligand (complex 8) | | Descriptor: | CARBON MONOXIDE, CHLORIDE ION, LYSOZYME C, ... | | Authors: | Santos, M.F.A, Mukhopadhyay, A, Romao, M.J, Romao, C.C, Santos-Silva, T. | | Deposit date: | 2014-08-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Contribution to the Rational Design of Ru(Co)3Cl2L Complexes for in Vivo Delivery of Co.
Dalton Trans, 44, 2015
|
|
7SFA
 
 | |
6RXG
 
 | |
6RY6
 
 | |
4PMT
 
 | |
4UZF
 
 | |
4V3H
 
 | | Crystal structure of CymA from Klebsiella oxytoca | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CYMA PROTEIN, NITRATE ION | | Authors: | van den Berg, B, Bhamidimarri, S.P, Kleinekathoefer, U, Winterhalter, M. | | Deposit date: | 2014-10-18 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Outer-membrane translocation of bulky small molecules by passive diffusion.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
4PNM
 
 | | Crystal Structure of human Tankyrase 2 in complex with Nu1025. | | Descriptor: | 1,2-ETHANEDIOL, 8-HYDROXY-2-METHYL-3-HYDRO-QUINAZOLIN-4-ONE, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UVM
 
 | | In meso crystal structure of the POT family transporter PepTSo | | Descriptor: | (2R)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLUTATHIONE UPTAKE TRANSPORTER | | Authors: | Lyons, J.A, Solcan, N, Caffrey, M, Newstead, S. | | Deposit date: | 2014-08-07 | | Release date: | 2015-02-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating Topology of the Proton-Coupled Oligopeptide Symporters.
Structure, 23, 2015
|
|
5IPC
 
 | | Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) H112N mutant nucleoside thiophosphoramidate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-[(S)-hydroxy{[2-(1H-indol-3-yl)ethyl]amino}phosphoryl]-5'-thioguanosine, CHLORIDE ION, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-03-09 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Caught before Released: Structural Mapping of the Reaction Trajectory for the Sofosbuvir Activating Enzyme, Human Histidine Triad Nucleotide Binding Protein 1 (hHint1).
Biochemistry, 56, 2017
|
|
6RZ7
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (F222 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
4PG5
 
 | |
6RZA
 
 | | Cryo-EM structure of the human inner arm dynein DNAH7 microtubule binding domain bound to microtubules | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1,Dynein heavy chain 7, axonemal,Cytoplasmic dynein 1 heavy chain 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
4PHQ
 
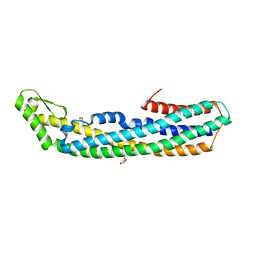 | | ClyA CC6/264 ox (6-303) | | Descriptor: | ACETATE ION, GLYCEROL, Hemolysin E, ... | | Authors: | Roderer, D.J.A, Glockshuber, R, Ban, N. | | Deposit date: | 2014-05-06 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Characterization of Variants of the Pore-Forming Toxin ClyA from Escherichia coli Controlled by a Redox Switch.
Biochemistry, 53, 2014
|
|
6RF2
 
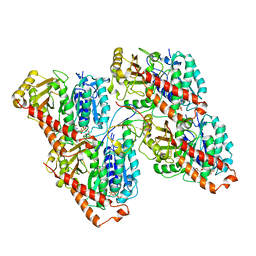 | |
4PIG
 
 | | Crystal structure of the ubiquitin K11S mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Loll, P.J, Xu, P.J, Schmidt, J, Melideo, S.L. | | Deposit date: | 2014-05-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Enhancing ubiquitin crystallization through surface-entropy reduction.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6RFN
 
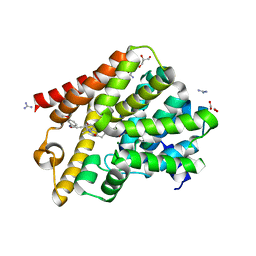 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1018 | | Descriptor: | 3-[5-[(4~{a}~{R},8~{a}~{S})-4-oxidanylidene-3-propan-2-yl-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-yl]-2-methoxy-phenyl]-~{N}-(3-methoxyphenyl)prop-2-ynamide, FORMIC ACID, GLYCEROL, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Alkynamide phthalazinones as a new class of TbrPDEB1 inhibitors (Part 2).
Bioorg.Med.Chem., 27, 2019
|
|
5I0M
 
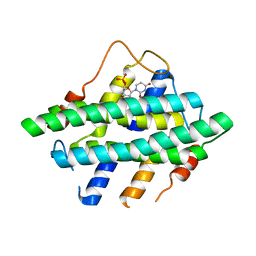 | |
5I1S
 
 | | Villin headpiece subdomain with a Lys30 to APC substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
4V2K
 
 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | Descriptor: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | Authors: | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-10-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
5I6Q
 
 | | Crystal structure of color device state B | | Descriptor: | DNA (26-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*GP*TP*GP*GP*TP*AP*TP*C)-3'), DNA (5'-D(*CP*AP*GP*AP*TP*AP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*TP*AP*CP*G)-3'), ... | | Authors: | Hao, Y, Birktoft, J, Seeman, N.C. | | Deposit date: | 2016-02-16 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.912 Å) | | Cite: | A device that operates within a self-assembled 3D DNA crystal.
Nat Chem, 9, 2017
|
|
6S5X
 
 | | Structure of RibR, the most N-terminal Rib domain from Group B Streptococcus species Streptococcus agalactiae | | Descriptor: | Group B streptococcal R4 surface protein, SODIUM ION | | Authors: | Whelan, F, Turkenburg, J.P, Griffiths, S.C, Bateman, A, Potts, J.R. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Defining the remarkable structural malleability of a bacterial surface protein Rib domain implicated in infection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4PKZ
 
 | |
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
