6D64
 
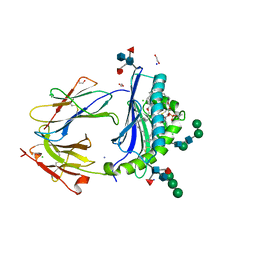 | | Crystal Structure of Human CD1b in Complex with POPC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-04-20 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
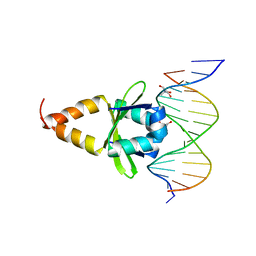
4ZM0
 
 | |
4ZG7
 
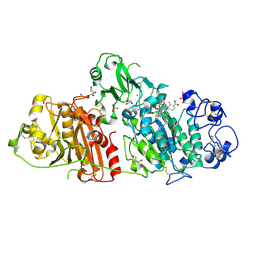 | | Structural basis for inhibition of human autotaxin by four novel compounds | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-({6-chloro-7-fluoro-2-methyl-1-[2-oxo-2-(spiro[cyclopropane-1,3'-indol]-1'(2'H)-yl)ethyl]-1H-indol-3-yl}sulfanyl)-2-fluorobenzoic acid, ... | | Authors: | Stein, A.J, Bain, G, Hutchinson, J.H, Evans, J.F. | | Deposit date: | 2015-04-22 | | Release date: | 2015-10-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibition of Human Autotaxin by Four Potent Compounds with Distinct Modes of Binding.
Mol.Pharmacol., 88, 2015
|
|
5PSH
 
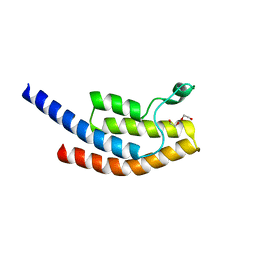 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 after initial refinement with no ligand modelled (structure 125) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, SODIUM ION | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
6DFG
 
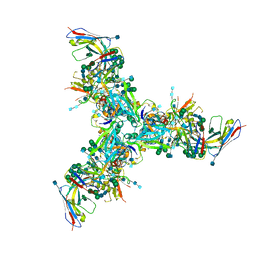 | | BG505 MD39 SOSIP trimer in complex with mature BG18 fragment antigen binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Ozorowski, G, Steichen, J.M, Schief, W.R, Ward, A.B. | | Deposit date: | 2018-05-14 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | A generalized HIV vaccine design strategy for priming of broadly neutralizing antibody responses.
Science, 366, 2019
|
|
1DTY
 
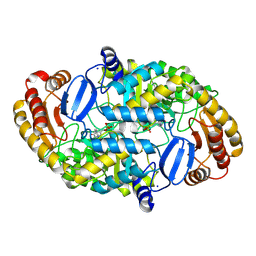 | | CRYSTAL STRUCTURE OF ADENOSYLMETHIONINE-8-AMINO-7-OXONANOATE AMINOTRANSFERASE WITH PYRIDOXAL PHOSPHATE COFACTOR. | | Descriptor: | ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION | | Authors: | Alexeev, D, Sawyer, L, Baxter, R.L, Alexeeva, M.V, Campopiano, D. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of adenosylmethionine-8-amino-7-oxonanoate aminotransferase with pyridoxal phosphate cofactor
to be published
|
|
5SCZ
 
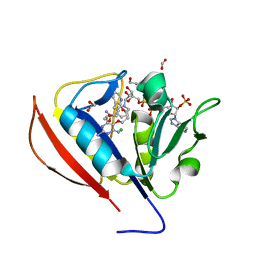 | |
6RFZ
 
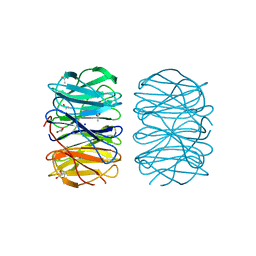 | | Photorhabdus laumondii lectin PLL2 in complex with D-glucose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SODIUM ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
6RG1
 
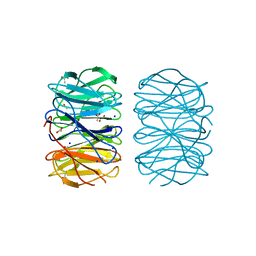 | | Photorhabdus laumondii lectin PLL2 in complex with L-rhamnose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SODIUM ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
5SCX
 
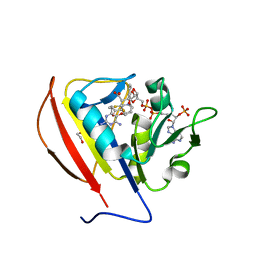 | |
6RG2
 
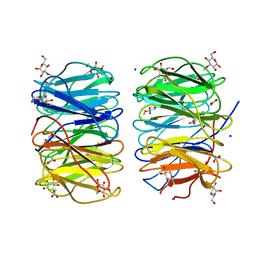 | | Photorhabdus laumondii lectin PLL2 in complex with 3-O-methyl-D-glucose | | Descriptor: | 3-O-methyl-alpha-D-glucopyranose, 3-O-methyl-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
6RGG
 
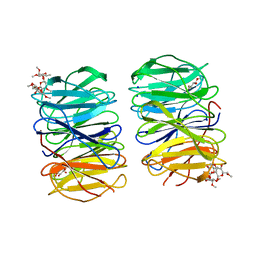 | | Photorhabdus laumondii lectin PLL2 in complex with O-methylated PGL-1-derived disaccharide | | Descriptor: | 1,2-ETHANEDIOL, 3,6-O-dimethyl-D-glucose, 6-deoxy-2,3-di-O-methyl-alpha-L-mannopyranose, ... | | Authors: | Houser, J, Fujdiarova, E, Wimmerova, M. | | Deposit date: | 2019-04-16 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heptabladed beta-propeller lectins PLL2 and PHL from Photorhabdus spp. recognize O-methylated sugars and influence the host immune system.
Febs J., 288, 2021
|
|
4Q56
 
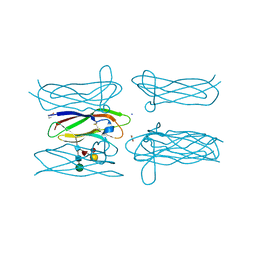 | | Structure of Helix aspersa agglutinin with natural glycosylation and N-acetyl-alpha-D-galactosamine (GalNAc) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, ACETATE ION, Helix aspersa agglutinin (HAA), ... | | Authors: | Pietrzyk, A.J, Bujacz, A, Bujacz, G. | | Deposit date: | 2014-04-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural studies of Helix aspersa agglutinin complexed with GalNAc: A lectin that serves as a diagnostic tool.
Int.J.Biol.Macromol., 81, 2015
|
|
5L2J
 
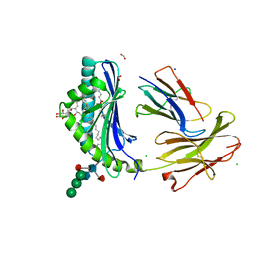 | | Crystal Structure of human CD1b in complex with C36-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, ACETATE ION, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
5L4O
 
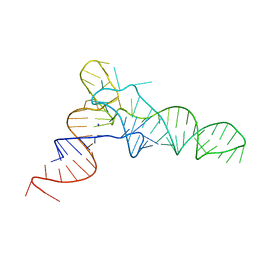 | |
6RU2
 
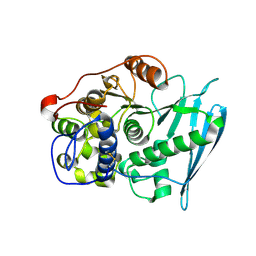 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-glucuronoyl methylesterase, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
5L2K
 
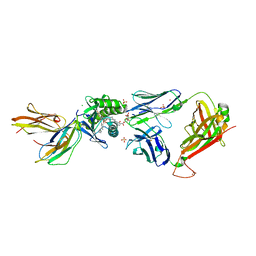 | | Crystal structure of GEM42 TCR-CD1b-GMM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, Beta-2-microglobulin, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
5L81
 
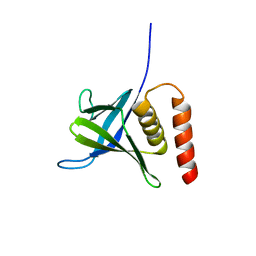 | |
5ELD
 
 | | Cholera toxin classical B-pentamer in complex with A Lewis-y | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cholera enterotoxin B subunit, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Heggelund, J.E, Burschowsky, D, Krengel, U. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Crystal Structures Elucidate the Molecular Basis of Cholera Blood Group Dependence.
Plos Pathog., 12, 2016
|
|
2YBF
 
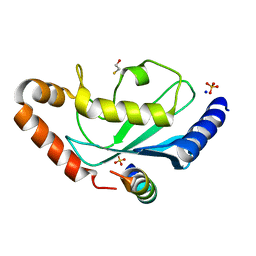 | | Complex of Rad18 (Rad6 binding domain) with Rad6b | | Descriptor: | BETA-MERCAPTOETHANOL, E3 UBIQUITIN-PROTEIN LIGASE RAD18, SODIUM ION, ... | | Authors: | Hibbert, R.G, Sixma, T.K. | | Deposit date: | 2011-03-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | E3 Ligase Rad18 Promotes Monoubiquitination Rather Than Ubiquitin Chain Formation by E2 Enzyme Rad6.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5LK0
 
 | |
6SEC
 
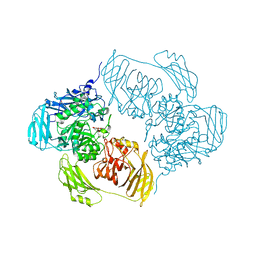 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG | | Descriptor: | 2-nitrophenyl beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.768 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
5LK1
 
 | |
6SEA
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in deep mode | | Descriptor: | ACETATE ION, Beta-galactosidase, SODIUM ION, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.869 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
6SE9
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E441Q in complex with lactose bound in shallow mode | | Descriptor: | ACETATE ION, Beta-galactosidase, FORMIC ACID, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.965 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
