1RTZ
 
 | |
1IUW
 
 | | P-HYDROXYBENZOATE HYDROXYLASE COMPLEXED WITH 4-4-HYDROXYBENZOATE AT PH 7.4 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOATE HYDROXYLASE, P-HYDROXYBENZOIC ACID | | Authors: | Gatti, D.L, Entsch, B, Ballou, D.P, Ludwig, M.L. | | Deposit date: | 1995-11-22 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis.
Biochemistry, 35, 1996
|
|
2H4Z
 
 | |
1QNL
 
 | |
2H2R
 
 | | Crystal structure of the human CD23 Lectin domain, apo form | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII)(Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
2H8I
 
 | |
2GSE
 
 | | Crystal Structure of Human Dihydropyrimidinease-like 2 | | Descriptor: | CALCIUM ION, Dihydropyrimidinase-related protein 2 | | Authors: | Ogg, D, Stenmark, P, Arrowsmith, C, Berglund, H, Collins, R, Edwards, A, Ehn, M, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Kotenyova, T, Kursula, P, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Holmberg-Schiavone, L, Thorsell, A.G, Uppenberg, J, Van Den Berg, S, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of human collapsin response mediator protein 2, a regulator of axonal growth.
J.Neurochem., 101, 2007
|
|
1HCY
 
 | |
5W0W
 
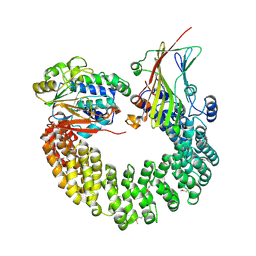 | | Crystal structure of Protein Phosphatase 2A bound to TIPRL | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, Serine/threonine-protein phosphatase 2A catalytic subunit alpha isoform, ... | | Authors: | Wu, C, Zheng, A, Li, J, Satyshur, K, Xing, Y. | | Deposit date: | 2017-06-01 | | Release date: | 2018-01-17 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Methylation-regulated decommissioning of multimeric PP2A complexes.
Nat Commun, 8, 2017
|
|
2H2T
 
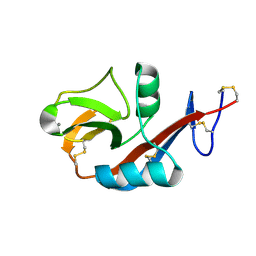 | | CD23 Lectin domain, Calcium 2+-bound | | Descriptor: | CALCIUM ION, Low affinity immunoglobulin epsilon Fc receptor (Lymphocyte IgE receptor) (Fc-epsilon-RII) (Immunoglobulin E-binding factor) (CD23 antigen) | | Authors: | Wurzburg, B.A. | | Deposit date: | 2006-05-19 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Changes in the Lectin Domain of CD23, the Low-Affinity IgE Receptor, upon Calcium Binding.
Structure, 14, 2006
|
|
2HU1
 
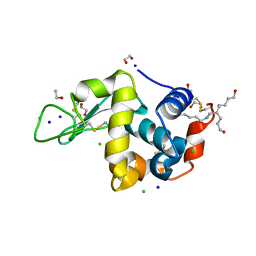 | | Crystal structure Analysis of Hen Egg White Lyszoyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wine, Y, Cohen-Hadar, N, Freeman, A, Lagziel-Simis, S, Frolow, F. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Elucidation of the mechanism and end products of glutaraldehyde crosslinking reaction by X-ray structure analysis
Biotechnol.Bioeng., 98, 2007
|
|
2HU7
 
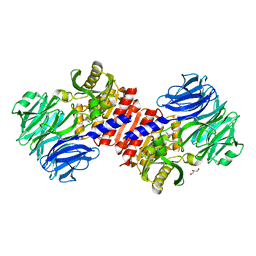 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | ACETYL GROUP, Acylamino-acid-releasing enzyme, GLYCEROL, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HHJ
 
 | | Human bisphosphoglycerate mutase complexed with 2,3-bisphosphoglycerate (15 days) | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 3-PHOSPHOGLYCERIC ACID, Bisphosphoglycerate mutase, ... | | Authors: | Wang, Y, Gong, W. | | Deposit date: | 2006-06-28 | | Release date: | 2006-10-24 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Seeing the process of histidine phosphorylation in human bisphosphoglycerate mutase.
J.Biol.Chem., 281, 2006
|
|
2HR1
 
 | |
2HU8
 
 | | Binding of inhibitors by Acylaminoacyl peptidase | | Descriptor: | 2-AMINOBENZOIC ACID, Acylamino-acid-releasing enzyme, GLYCINE, ... | | Authors: | Kiss, A.L, Hornung, B, Radi, K, Gengeliczki, Z, Sztaray, B, Harmat, V, Polgar, L. | | Deposit date: | 2006-07-26 | | Release date: | 2007-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity
J.Mol.Biol., 368, 2007
|
|
2HJB
 
 | |
1SGT
 
 | |
2HSD
 
 | |
2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
2HPA
 
 | | STRUCTURAL ORIGINS OF L(+)-TARTRATE INHIBITION OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-PROPYL-TARTRAMIC ACID, ... | | Authors: | Lacount, M.W, Handy, G, Lebioda, L. | | Deposit date: | 1998-09-11 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural origins of L(+)-tartrate inhibition of human prostatic acid phosphatase.
J.Biol.Chem., 273, 1998
|
|
2FY7
 
 | | Crystal structure of the catalytic domain of the human beta1,4-galactosyltransferase mutant M339H in apo form | | Descriptor: | Beta-1,4-galactosyltransferase 1, TRIETHYLENE GLYCOL | | Authors: | Ramakrishnan, B, Ramasamy, V, Qasba, P.K. | | Deposit date: | 2006-02-07 | | Release date: | 2006-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Snapshots of beta-1,4-Galactosyltransferase-I Along the Kinetic Pathway.
J.Mol.Biol., 357, 2006
|
|
2H94
 
 | |
2H4T
 
 | | Crystal structure of rat carnitine palmitoyltransferase II | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial, DODECANE | | Authors: | Hsiao, Y.S, Jogl, G, Esser, V, Tong, L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rat carnitine palmitoyltransferase II (CPT-II).
Biochem.Biophys.Res.Commun., 346, 2006
|
|
2H65
 
 | | Crystal strusture of caspase-3 with inhibitor Ac-VDVAD-Cho | | Descriptor: | Ac-VDVAD-Cho, caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-30 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
2GMT
 
 | | THREE-DIMENSIONAL STRUCTURE OF CHYMOTRYPSIN INACTIVATED WITH (2S) N-ACETYL-L-ALANYL-L-PHENYLALANYL-CHLOROETHYL KETONE: IMPLICATIONS FOR THE MECHANISM OF INACTIVATION OF SERINE PROTEASES BY CHLOROKETONES | | Descriptor: | (2S) N-ACETYL-L-ALANYL-ALPHAL-PHENYLALANYL-CHLOROETHYLKETONE, GAMMA-CHYMOTRYPSIN | | Authors: | Kreutter, K, Steinmetz, A.C.U, Liang, T.-C, Prorok, M, Abeles, R, Ringe, D. | | Deposit date: | 1994-09-07 | | Release date: | 1994-11-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of chymotrypsin inactivated with (2S)-N-acetyl-L-alanyl-L-phenylalanyl alpha-chloroethane: implications for the mechanism of inactivation of serine proteases by chloroketones.
Biochemistry, 33, 1994
|
|
