2HP1
 
 | | Inter-subunit signaling in GSAM | | Descriptor: | (4S)-4,5-DIAMINOPENTANOIC ACID, (4S)-4-AMINO-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]PENTANOIC ACID, Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) plp-form, ... | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HP2
 
 | | Inter-subunit signaling in GSAM | | Descriptor: | (4R)-5-AMINO-4-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]PENTANOIC ACID, (4S)-4,5-DIAMINOPENTANOIC ACID, Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) hybrid-form, ... | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HOZ
 
 | | Inter-subunit signaling in GSAM | | Descriptor: | (4S)-4,5-DIAMINOPENTANOIC ACID, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) pmp-form | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HOY
 
 | | Inter-subunit signaling in GSAM | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase (GSAM) apo-form, PHOSPHATE ION | | Authors: | Stetefeld, J. | | Deposit date: | 2006-07-17 | | Release date: | 2006-08-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Intersubunit signaling in glutamate-1-semialdehyde-aminomutase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4JTH
 
 | | Crystal structure of F114R/R117Q mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
2KA1
 
 | |
4JTE
 
 | | Crystal structure of F114A mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, SODIUM ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTF
 
 | | Crystal structure of F114R mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTG
 
 | | Crystal structure of F114R/R117A mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
4JTK
 
 | | Crystal structure of R117Q mutant of 3-deoxy-D-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, SODIUM ION | | Authors: | Allison, T.M, Cochrane, F.C, Jameson, G.B, Parker, E.J. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Examining the Role of Intersubunit Contacts in Catalysis by 3-Deoxy-d-manno-octulosonate 8-Phosphate Synthase.
Biochemistry, 52, 2013
|
|
2JYG
 
 | |
2L1R
 
 | | The structure of the calcium-sensitizer, dfbp-o, in complex with the N-domain of troponin C and the switch region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Robertson, I.M, Sun, Y, Li, M.X, Sykes, B.D. | | Deposit date: | 2010-08-03 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A structural and functional perspective into the mechanism of Ca(2+)-sensitizers that target the cardiac troponin complex.
J.MOL.CELL.CARDIOL., 49, 2010
|
|
8BGF
 
 | |
8HC0
 
 | | Crystal structure of the extracellular domains of GPR110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Adhesion G-protein coupled receptor F1, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wang, F.F, Song, G.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Extracellular Domains of GPR110.
J.Mol.Biol., 435, 2023
|
|
8EEG
 
 | |
8EEM
 
 | |
8H4M
 
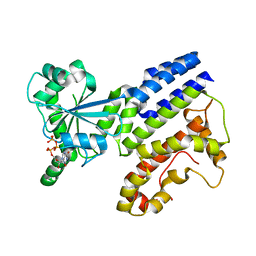 | | Crystal Structure of GTP-bound Irgb6_T95D mutant | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, T-cell-specific guanine nucleotide triphosphate-binding protein 2 | | Authors: | Saijo-Hamano, Y, Okuma, H, Sakai, N, Kato, T, Imasaki, T, Nitta, R. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis of Irgb6 inactivation by Toxoplasma gondii through the phosphorylation of switch I
To Be Published
|
|
8EEH
 
 | |
8EEO
 
 | |
8EEF
 
 | |
8EEN
 
 | |
8EEI
 
 | |
5LDW
 
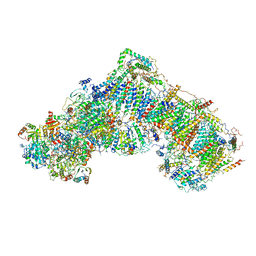 | | Structure of mammalian respiratory Complex I, class1 | | Descriptor: | Acyl carrier protein, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Vinothkumar, K.R, Zhu, J, Hirst, J. | | Deposit date: | 2016-06-28 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structure of mammalian respiratory complex I.
Nature, 536, 2016
|
|
8EEJ
 
 | |
8EEK
 
 | |
