5XX4
 
 | | A BPTI-[5,55] variant with C14GA38K mutations | | Descriptor: | Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M. | | Deposit date: | 2017-07-01 | | Release date: | 2018-07-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrophobic surface residues can stabilize a protein through improved water-protein interactions.
Febs J., 2019
|
|
2KXW
 
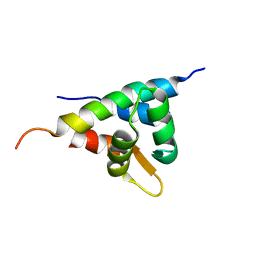 | |
2KPY
 
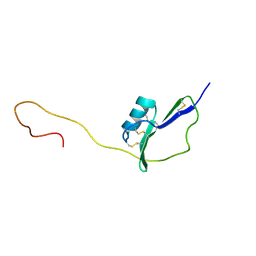 | | Solution Structure of the major allergen of Artemisia vulgaris (Art v 1) | | Descriptor: | Major pollen allergen Art v 1 | | Authors: | Razzera, G, Gadermaier, G, Almeida, M, Egger, M, Jahn-Schmid, B, Almeida, F, Ferreira, F, Valente, A. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mapping the interactions between a major pollen allergen and human IgE antibodies.
Structure, 18, 2010
|
|
5UXY
 
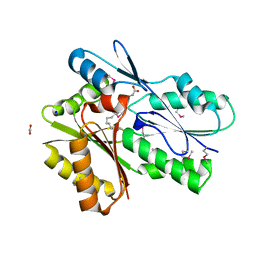 | | The crystal structure of a DegV family protein from Eubacterium eligens loaded with heptadecanoic acid to 1.80 Angstrom resolution (ALTERNATIVE REFINEMENT OF PDB 3FDJ with HEPTADECANOIC acid) | | Descriptor: | ACETIC ACID, DegV family protein, SODIUM ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-02-23 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
6DWV
 
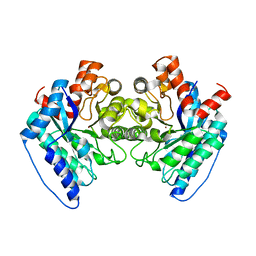 | | Crystal structure of the LigJ Hydratase in the Apo state | | Descriptor: | 4-oxalomesaconate hydratase, ZINC ION | | Authors: | Mabanglo, M.F, Raushel, F.M. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Reaction Mechanism of the LigJ Hydratase: An Enzyme Critical for the Bacterial Degradation of Lignin in the Protocatechuate 4,5-Cleavage Pathway.
Biochemistry, 57, 2018
|
|
3OMF
 
 | |
3VM7
 
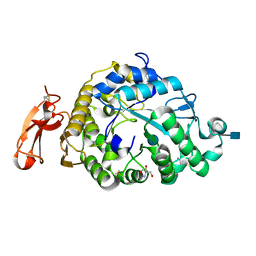 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
1L9T
 
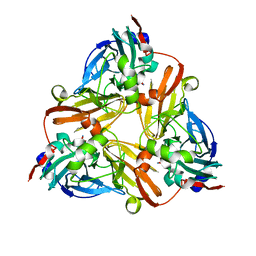 | |
6A9V
 
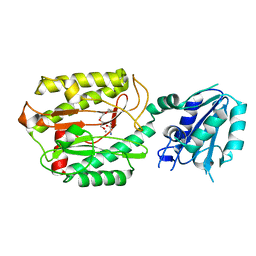 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 42 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
6A9T
 
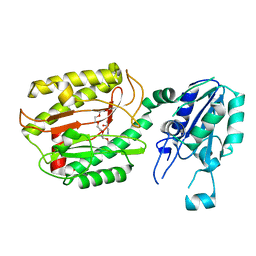 | | Crystal structure of Icp55 from Saccharomyces cerevisiae (N-terminal 58 residues deletion) | | Descriptor: | GLYCINE, Intermediate cleaving peptidase 55, MANGANESE (II) ION, ... | | Authors: | Singh, R, Kumar, A, Goyal, V.D, Makde, R.D. | | Deposit date: | 2018-07-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures and biochemical analyses of intermediate cleavage peptidase: role of dynamics in enzymatic function.
FEBS Lett., 593, 2019
|
|
1VIU
 
 | |
1VIZ
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | PcrB protein homolog, SODIUM ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
5UK9
 
 | | Wild-type K-Ras(GCP) pH 6.5 | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Parker, J.A, Mattos, C. | | Deposit date: | 2017-01-20 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | K-Ras Populates Conformational States Differently from Its Isoform H-Ras and Oncogenic Mutant K-RasG12D.
Structure, 26, 2018
|
|
1L9P
 
 | |
1L9S
 
 | |
5MJG
 
 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
1O6B
 
 | |
1VHD
 
 | |
5V85
 
 | | The crystal structure of the protein of DegV family COG1307 from Ruminococcus gnavus ATCC 29149 (alternative refinement of PDB 3JR7 with Vaccenic acid) | | Descriptor: | EDD domain protein, DegV family, PHOSPHATE ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-03-21 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
1W6B
 
 | | Solution NMR Structure of a Long Neurotoxin from the Venom of the Asian Cobra, 20 Structures | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Talebzadeh-Farooji, M, Amininasab, M, Elmi, M.M, Naderi-Manesh, H, Sarbolouki, M.N. | | Deposit date: | 2004-08-17 | | Release date: | 2004-12-22 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of long neurotoxin NTX-1 from the venom of Naja naja oxiana by 2D-NMR spectroscopy.
Eur. J. Biochem., 271, 2004
|
|
1VH0
 
 | | Crystal structure of a hypothetical protein | | Descriptor: | Hypothetical UPF0247 protein SAV0024/SA0023 | | Authors: | Structural GenomiX | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
5LIV
 
 | | Crystal structure of myxobacterial CYP260A1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome P450 CYP260A1,Cytochrome P450 CYP260A1, DIMETHYL SULFOXIDE, ... | | Authors: | Carius, Y, Khatri, Y, Bernhardt, R, Lancaster, C.R.D. | | Deposit date: | 2016-07-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of CYP260A1 from Sorangium cellulosum to investigate the 1 alpha-hydroxylation of a mineralocorticoid.
FEBS Lett., 590, 2016
|
|
1PD8
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine Antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PD9
 
 | | Analysis of Three Crystal Structure Determinations of a 5-Methyl-6-N-Methylanilino Pyridopyrimidine antifolate Complex with Human Dihydrofolate Reductase | | Descriptor: | 2,4-DIAMINO-5-METHYL-6-[(3,4,5-TRIMETHOXY-N-METHYLANILINO)METHYL]PYRIDO[2,3-D]PYRIMIDINE, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Luft, J.R, Pangborn, W, Gangjee, A. | | Deposit date: | 2003-05-19 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of three crystal structure determinations of a 5-methyl-6-N-methylanilino pyridopyrimidine antifolate complex with human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5Z7B
 
 | |
