2BT2
 
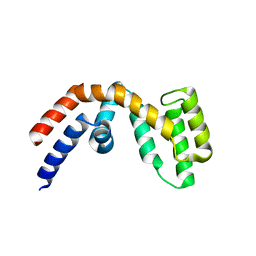 | | Structure of the regulator of G-protein signaling 16 | | Descriptor: | REGULATOR OF G-PROTEIN SIGNALING 16 | | Authors: | Bunkoczi, G, Haroniti, A, Longman, E, Niesen, F, Soundararajan, M, Ball, L.J, von Delft, F, Doyle, D.A, Arrowsmith, C, Edwards, A, Sundstrom, M. | | Deposit date: | 2005-05-25 | | Release date: | 2005-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Diversity in the Rgs Domain and its Interaction with Heterotrimeric G Protein Alpha- Subunits.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
8B07
 
 | |
2WHN
 
 | |
2WQ1
 
 | |
7M1H
 
 | |
7LZP
 
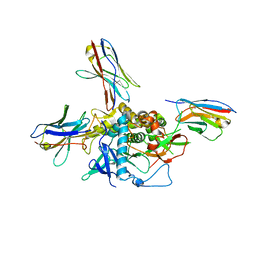 | | LC/A-JPU-B9-JPU-A11-JPU-G11 | | Descriptor: | Botulinum neurotoxin A light chain, JPU-A11, JPU-B9, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
8AFQ
 
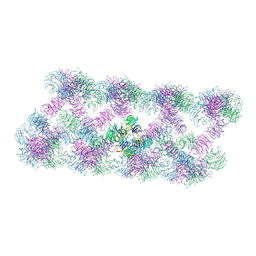 | | Tube assembly of Atg18-PR72AA | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFW
 
 | | Tube assembly of Atg18-WT | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8AFY
 
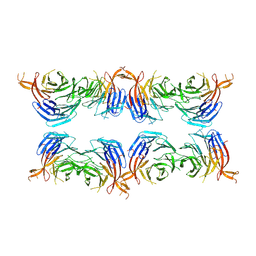 | | Subtomogram average of membrane-bound Atg18 oligomers | | Descriptor: | Autophagy-related protein 18 | | Authors: | Mann, D, Fromm, S, Martinez-Sanchez, A, Gopaldass, N, Mayer, A, Sachse, C. | | Deposit date: | 2022-07-18 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Atg18 oligomer organization in assembled tubes and on lipid membrane scaffolds.
Nat Commun, 14, 2023
|
|
8ABY
 
 | |
8AC0
 
 | |
8AD1
 
 | |
8ACP
 
 | |
8ABZ
 
 | |
8AC2
 
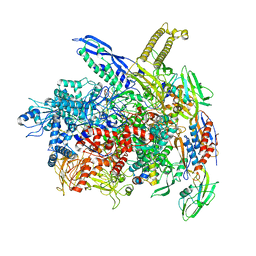 | | RNA polymerase- post-terminated, open clamp state | | Descriptor: | DNA Non-template strand, DNA Template strand, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Dey, S, Weixlbaumer, A. | | Deposit date: | 2022-07-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into RNA-mediated transcription regulation in bacteria.
Mol.Cell, 82, 2022
|
|
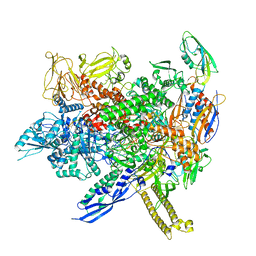
8AC1
 
 | |
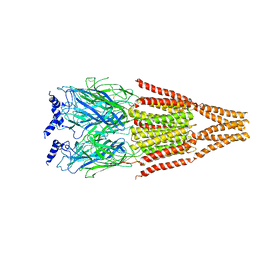
2BG9
 
 | |
8AYS
 
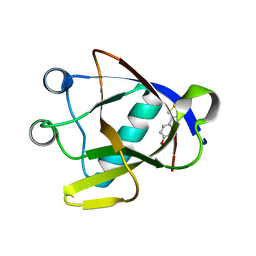 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8AZ8
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
2WQ2
 
 | |
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2CCE
 
 | | Parallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
2CMY
 
 | | Crystal complex between bovine trypsin and Veronica hederifolia trypsin inhibitor | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, GLYCEROL, ... | | Authors: | Conners, R, Yardley, J.L, Konarev, A, Shewry, P, Brady, R.L. | | Deposit date: | 2006-05-16 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An Unusual Helix-Turn-Helix Protease Inhibitory Motif in a Novel Trypsin Inhibitor from Seeds of Veronica (Veronica Hederifolia L.).
J.Biol.Chem., 282, 2007
|
|
2CCF
 
 | | Antiparallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
7ZIU
 
 | |
