1BZD
 
 | |
1BZE
 
 | |
1FEH
 
 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
1BZA
 
 | | BETA-LACTAMASE TOHO-1 FROM ESCHERICHIA COLI TUH12191 | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Ibuka, A, Taguchi, A, Ishiguro, M, Fushinobu, S, Ishii, Y, Kamitori, S, Okuyama, K, Yamaguchi, K, Konno, M, Matsuzawa, H. | | Deposit date: | 1998-10-28 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the E166A mutant of extended-spectrum beta-lactamase Toho-1 at 1.8 A resolution.
J.Mol.Biol., 285, 1999
|
|
1BZF
 
 | | NMR SOLUTION STRUCTURE AND DYNAMICS OF THE COMPLEX OF LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE WITH THE NEW LIPOPHILIC ANTIFOLATE DRUG TRIMETREXATE, 22 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, TRIMETREXATE | | Authors: | Polshakov, V.I, Birdsall, B, Frenkiel, T.A, Gargaro, A.R, Feeney, J. | | Deposit date: | 1998-10-28 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics in solution of the complex of Lactobacillus casei dihydrofolate reductase with the new lipophilic antifolate drug trimetrexate.
Protein Sci., 8, 1999
|
|
1DCN
 
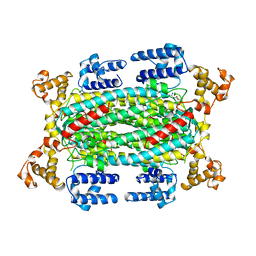 | | INACTIVE MUTANT H162N OF DELTA 2 CRYSTALLIN WITH BOUND ARGININOSUCCINATE | | Descriptor: | ARGININOSUCCINATE, DELTA 2 CRYSTALLIN | | Authors: | Vallee, F, Turner, M.A, Lindley, P, Howell, P.L. | | Deposit date: | 1998-10-29 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an inactive duck delta II crystallin mutant with bound argininosuccinate.
Biochemistry, 38, 1999
|
|
1TUX
 
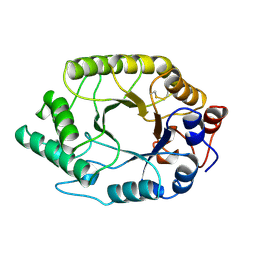 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS | | Descriptor: | XYLANASE | | Authors: | Natesh, R, Bhanumoorthy, P, Vithayathil, P.J, Sekar, K, Ramakumar, S, Viswamitra, M.A. | | Deposit date: | 1998-10-29 | | Release date: | 1999-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and proposed amino acid sequence of a thermostable xylanase from Thermoascus aurantiacus.
J.Mol.Biol., 288, 1999
|
|
1BZJ
 
 | | Human ptp1b complexed with tpicooh | | Descriptor: | 6-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALENE-2-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE) | | Authors: | Groves, M.R, Yao, Z.-J, Barford Jr, D.T.B. | | Deposit date: | 1998-10-29 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
1RNF
 
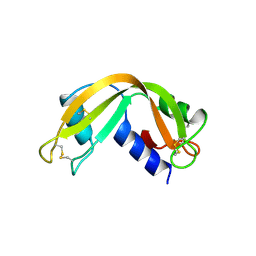 | | X-RAY CRYSTAL STRUCTURE OF UNLIGANDED HUMAN RIBONUCLEASE 4 | | Descriptor: | PROTEIN (RIBONUCLEASE 4) | | Authors: | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 1998-10-29 | | Release date: | 1999-10-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
5TLI
 
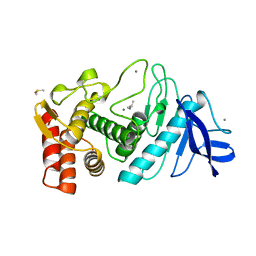 | | THERMOLYSIN (60% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
6TLI
 
 | | THERMOLYSIN (60% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
4TLI
 
 | | THERMOLYSIN (25% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-29 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
2NMB
 
 | | DNUMB PTB DOMAIN COMPLEXED WITH A PHOSPHOTYROSINE PEPTIDE, NMR, ENSEMBLE OF STRUCTURES. | | Descriptor: | PROTEIN (GPPY PEPTIDE), PROTEIN (NUMB PROTEIN) | | Authors: | Li, S.-C, Zwahlen, C, Vincent, S.J.F, McGlade, C.J, Pawson, T, Forman-Kay, J.D. | | Deposit date: | 1998-10-29 | | Release date: | 1998-11-04 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Numb PTB domain-peptide complex suggests a basis for diverse binding specificity.
Nat.Struct.Biol., 5, 1998
|
|
8TLI
 
 | | THERMOLYSIN (100% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
7TLI
 
 | | THERMOLYSIN (90% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
2UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1UUG
 
 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1BZK
 
 | | STRUCTURAL STUDIES ON THE EFFECTS OF THE DELETION IN THE RED CELL ANION EXCHANGER (BAND3, AE1) ASSOCIATED WITH SOUTH EAST ASIAN OVALOCYTOSIS. | | Descriptor: | PROTEIN (BAND 3 ANION TRANSPORT PROTEIN) | | Authors: | Chambers, E.J, Bloomberg, G.B, Ring, S.M, Tanner, M.J.A. | | Deposit date: | 1998-11-01 | | Release date: | 1999-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the effects of the deletion in the red cell anion exchanger (band 3, AE1) associated with South East Asian ovalocytosis.
J.Mol.Biol., 285, 1999
|
|
2PVI
 
 | | PVUII ENDONUCLEASE COMPLEXED TO AN IODINATED COGNATE DNA | | Descriptor: | DNA (5'-D(*TP*GP*AP*CP*CP*AP*GP*(C38)P*TP*GP*GP*TP*C)-3'), TYPE II RESTRICTION ENZYME PVUII | | Authors: | Horton, J, Cheng, X. | | Deposit date: | 1998-11-01 | | Release date: | 1999-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | How is modification of the DNA substrate recognized by the PvuII restriction endonuclease?
J.Biol.Chem., 379, 1998
|
|
1TLX
 
 | | THERMOLYSIN (NATIVE) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-11-02 | | Release date: | 2000-03-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
2SEM
 
 | | SEM5 SH3 DOMAIN COMPLEXED WITH PEPTOID INHIBITOR | | Descriptor: | PROTEIN (SEX MUSCLE ABNORMAL PROTEIN 5), PROTEIN (SH3 PEPTOID INHIBITOR) | | Authors: | Nguyen, J.T, Turck, C.W, Cohen, F.E, Zuckermann, R.N, Lim, W.A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors.
Science, 282, 1998
|
|
1BZL
 
 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH TRYPANOTHIONE, AND THE STRUCTURE-BASED DISCOVERY OF NEW NATURAL PRODUCT INHIBITORS | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) | | Authors: | Bond, C.S, Zhang, Y, Berriman, M, Cunningham, M, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 1998-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Trypanosoma cruzi trypanothione reductase in complex with trypanothione, and the structure-based discovery of new natural product inhibitors.
Structure Fold.Des., 7, 1999
|
|
1BZO
 
 | | THREE-DIMENSIONAL STRUCTURE OF PROKARYOTIC CU,ZN SUPEROXIDE DISMUTASE FROM P.LEIOGNATHI, SOLVED BY X-RAY CRYSTALLOGRAPHY. | | Descriptor: | COPPER (II) ION, PROTEIN (SUPEROXIDE DISMUTASE), URANYL (VI) ION, ... | | Authors: | Bordo, D, Matak, D, Djinovic-Carugo, K, Rosano, C, Pesce, A, Bolognesi, M, Stroppolo, M.E, Falconi, M, Battistoni, A, Desideri, A. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolutionary constraints for dimer formation in prokaryotic Cu,Zn superoxide dismutase.
J.Mol.Biol., 285, 1999
|
|
1BZP
 
 | |
1P32
 
 | | CRYSTAL STRUCTURE OF HUMAN P32, A DOUGHNUT-SHAPED ACIDIC MITOCHONDRIAL MATRIX PROTEIN | | Descriptor: | MITOCHONDRIAL MATRIX PROTEIN, SF2P32 | | Authors: | Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 1998-11-02 | | Release date: | 1999-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
