8RCS
 
 | |
8CAD
 
 | |
8CA9
 
 | |
8QEY
 
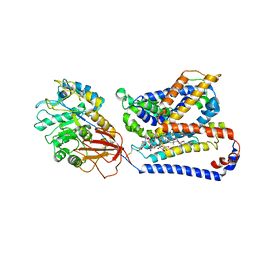 | | Structure of human Asc1/CD98hc heteromeric amino acid transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Martinez-Molledo, M, Rullo-Tubau, J, Errasti-Murugarren, E, Palacin, M, Llorca, O. | | Deposit date: | 2023-09-01 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and mechanisms of transport of human Asc1/CD98hc amino acid transporter.
Nat Commun, 15, 2024
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8CDD
 
 | |
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8R3V
 
 | |
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8CGR
 
 | | Apramycin bound to the 30S body | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S16, ... | | Authors: | Paternoga, H, Koller, T.O, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8QO9
 
 | | Cryo-EM structure of a human spliceosomal B complex protomer | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-09-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (5.29 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
1PTF
 
 | |
1Q61
 
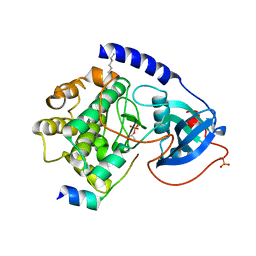 | | PKA triple mutant model of PKB | | Descriptor: | N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1PX2
 
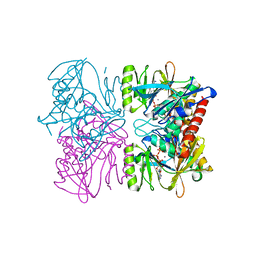 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP (Form 1) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Synapsin I | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-07-02 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
8R94
 
 | |
8R9L
 
 | |
8RAA
 
 | |
2VCP
 
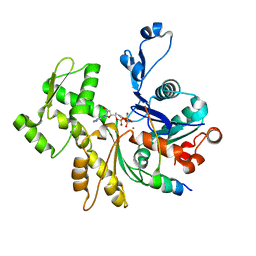 | | Crystal structure of N-Wasp VC domain in complex with skeletal actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gaucher, J.F, Didry, D, Carlier, M.F. | | Deposit date: | 2007-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Interactions of isolated C-terminal fragments of neural Wiskott-Aldrich syndrome protein (N-WASP) with actin and Arp2/3 complex.
J. Biol. Chem., 287, 2012
|
|
8R8F
 
 | |
8R90
 
 | |
8R8Z
 
 | |
