5DUL
 
 | | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Osipiuk, J, Mulligan, R, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Yersinia pestis in complex with NADPH .
to be published
|
|
4ZN6
 
 | |
6MCR
 
 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | Authors: | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | Deposit date: | 2018-09-02 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
7C5N
 
 | | Crystal Structure of C150A+H177A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli at 2.0 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5I
 
 | | Crystal Structure of C150A mutant of Glyceraldehyde-3-phosphate-dehydrogenase1 from Escherichia coli complexed with PO4 at 2.49 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
7C5R
 
 | | Crystal Structure of C150S mutant Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli complexed with BPG at 2.31 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
8R41
 
 | | Structure of CHI3L1 in complex with inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Nowak, E, Napiorkowska-Gromadzka, A, Nowotny, M. | | Deposit date: | 2023-11-10 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Discovery of High-Affinity Small Molecule Ligands and Development of Tool Probes to Study the Role of Chitinase-3-Like Protein 1.
J.Med.Chem., 67, 2024
|
|
5DUF
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK729A | | Descriptor: | (5R,7S)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxylic acid, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5HBM
 
 | | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2015-12-31 | | Release date: | 2016-02-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.043 Å) | | Cite: | Crystal Structure of a Dihydroxycoumarin RNase H Active-Site Inhibitor in Complex with HIV-1 Reverse Transcriptase
To Be Published
|
|
6USW
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
5EQV
 
 | | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-MALATE, FE (III) ION, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 Angstrom Crystal Structure of Bifunctional 2',3'-cyclic Nucleotide 2'-phosphodiesterase/3'-Nucleotidase Periplasmic Precursor Protein from Yersinia pestis with Phosphate bound to the Active site.
To Be Published
|
|
6JO3
 
 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7LOO
 
 | | S-adenosyl methionine transferase cocrystallized with ATP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Jackson, C.J, Tan, L.L, Laurino, P. | | Deposit date: | 2021-02-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
5OOI
 
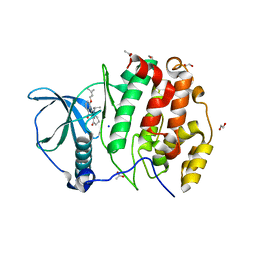 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5T8K
 
 | | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD.
To Be Published
|
|
6AYV
 
 | | Crystal structure of fructose-1,6-bisphosphatase T84A from Mycobacterium tuberculosis | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase class 2, GLYCEROL, ... | | Authors: | Abad-Zapatero, C, Wolf, N, Gutka, H.J, Movahedzadeh, F. | | Deposit date: | 2017-09-08 | | Release date: | 2018-03-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Mycobacterium tuberculosis GlpX protein (class II fructose-1,6-bisphosphatase): implications for the active oligomeric state, catalytic mechanism and citrate inhibition.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7C5H
 
 | | Crystal Structure of Glyceraldehyde-3-phosphate dehydrogenase1 from Escherichia coli at 2.09 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L, Liu, M.R, Bao, L.Y, Yao, Y.C, Bostrom, I.K, Wang, Y.D, Chen, A.Q, Li, J.X, Gu, S.H, Ji, C.N. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Novel Structures of Type 1 Glyceraldehyde-3-phosphate Dehydrogenase from Escherichia coli Provide New Insights into the Mechanism of Generation of 1,3-Bisphosphoglyceric Acid.
Biomolecules, 11, 2021
|
|
6HMG
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-(1,2-anhydro-carba-glucosamine) | | Descriptor: | (1~{S},2~{R},3~{R},4~{R},6~{S})-4-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3-diol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Lu, D, Zhu, S, Bernardo-Seisdedos, G, Millet, O, Zhang, Y, Sollogoub, M, Jimenez-Barbero, J, Davies, G.J. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | From 1,4-Disaccharide to 1,3-Glycosyl Carbasugar: Synthesis of a Bespoke Inhibitor of Family GH99 Endo-alpha-mannosidase.
Org.Lett., 20, 2018
|
|
3K2E
 
 | |
5HP1
 
 | |
6MK9
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl {(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease | | Authors: | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-121
To Be Published
|
|
8ING
 
 | | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmad, M.I, Viswanathan, V, Kumar, M, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution
To be published
|
|
5DUC
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK951A | | Descriptor: | (5R,7S)-N-(1,3-benzodioxol-5-ylmethyl)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | Authors: | Cox, J.A.G, Besra, G.S, Futterer, K. | | Deposit date: | 2015-09-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5UMH
 
 | |
