5LM4
 
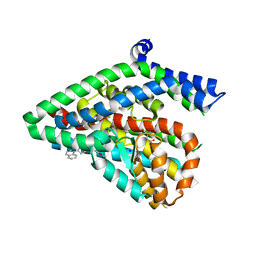 | | Structure of the thermostalilized EAAT1 cryst-II mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
7L4E
 
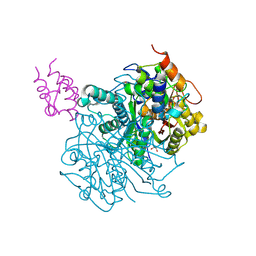 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabF, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] synthase 2, Acyl carrier protein, ... | | Authors: | Mindrebo, J.T, Chen, A, Kim, W.E, Burkart, M.D, Noel, J.P. | | Deposit date: | 2020-12-18 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Mechanistic Analyses of the Gating Mechanism of Elongating Ketosynthases
Acs Catalysis, 11, 2021
|
|
1J5M
 
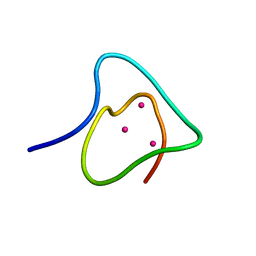 | | SOLUTION STRUCTURE OF THE SYNTHETIC 113CD_3 BETA_N DOMAIN OF LOBSTER METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Munoz, A, Forsterling, F.H, Shaw III, C.F, Petering, D.H. | | Deposit date: | 2002-05-16 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the (113)Cd(3)beta domains from Homarus americanus metallothionein-1: hydrogen bonding and solvent accessibility of sulfur atoms
J.Biol.Inorg.Chem., 7, 2002
|
|
1JCJ
 
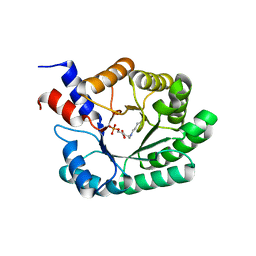 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1JK7
 
 | | CRYSTAL STRUCTURE OF THE TUMOR-PROMOTER OKADAIC ACID BOUND TO PROTEIN PHOSPHATASE-1 | | Descriptor: | BETA-MERCAPTOETHANOL, MANGANESE (II) ION, OKADAIC ACID, ... | | Authors: | Maynes, J.T, Bateman, K.S, Cherney, M.M, Das, A.K, James, M.N. | | Deposit date: | 2001-07-11 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the tumor-promoter okadaic acid bound to protein phosphatase-1.
J.Biol.Chem., 276, 2001
|
|
7L1G
 
 | | PRMT5-MEP50 Complexed with SAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Development of a Flexible and Robust Synthesis of Tetrahydrofuro[3,4- b ]furan Nucleoside Analogues.
J.Org.Chem., 86, 2021
|
|
4W9O
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1R)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1IF1
 
 | | INTERFERON REGULATORY FACTOR 1 (IRF-1) COMPLEX WITH DNA | | Descriptor: | DNA (26-MER), PROTEIN (INTERFERON REGULATORY FACTOR 1) | | Authors: | Escalante, C.R, Yie, J, Thanos, D, Aggarwal, A. | | Deposit date: | 1997-09-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of IRF-1 with bound DNA reveals determinants of interferon regulation.
Nature, 391, 1998
|
|
8BV9
 
 | | Acylphosphatase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, GLYCEROL, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First 3-D structural evidence of a native-like intertwined dimer in the acylphosphatase family.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
4WCF
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 9 | | Descriptor: | 3-(5-amino-1,3,4-thiadiazol-2-yl)pyridin-4-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
7KWA
 
 | | Structure of DCN1 bound to N-((4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-(trifluoromethyl)benzamide | | Descriptor: | Endolysin,DCN1-like protein 1, N-[(4S,5S)-3-(aminomethyl)-7-ethyl-4-(4-fluorophenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-(trifluoromethyl)benzamide | | Authors: | Kim, H.S, Hammill, J.T, Schulman, B.A, Guy, R.K, Scott, D.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Improvement of Oral Bioavailability of Pyrazolo-Pyridone Inhibitors of the Interaction of DCN1/2 and UBE2M.
J.Med.Chem., 64, 2021
|
|
5M03
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-noeuromycin and 1,2-alpha-mannobiose | | Descriptor: | (2S,3S,4R,5R)-2,3,4-TRIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
4W9P
 
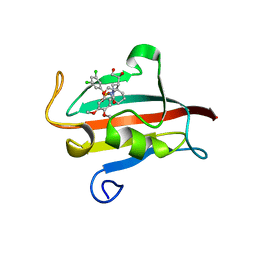 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1IYP
 
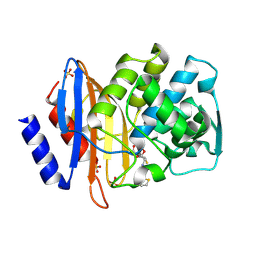 | | Toho-1 beta-Lactamase In Complex With Cephalothin | | Descriptor: | CEPHALOTHIN GROUP, SULFATE ION, Toho-1 beta-lactamase | | Authors: | Shimamura, T, Ibuka, A, Fushinobu, S, Wakagi, T, Ishiguro, M, Ishii, Y, Matsuzawa, H. | | Deposit date: | 2002-09-04 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acyl-intermediate Structures of the Extended-spectrum Class A beta -Lactamase, Toho-1, in Complex with Cefotaxime, Cephalothin, and Benzylpenicillin.
J.Biol.Chem., 277, 2002
|
|
6P8Q
 
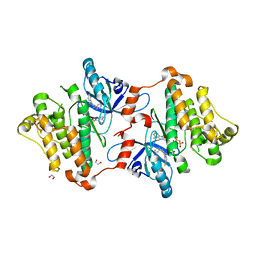 | | EGFR in complex with a dihydrodibenzodiazepinone allosteric inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 10-benzyl-2-fluoro-5,10-dihydro-11H-dibenzo[b,e][1,4]diazepin-11-one, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Yun, C.H, Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Optimization of Dibenzodiazepinones as Allosteric Mutant-Selective EGFR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7KV0
 
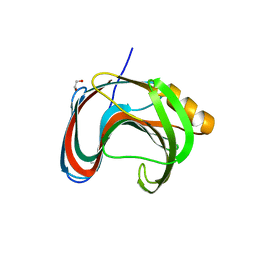 | | Crystallographic structure of Paenibacillus xylanivorans GH11 | | Descriptor: | 1,2-ETHANEDIOL, Endo-1,4-beta-xylanase | | Authors: | Briganti, L, Polikarpov, I. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and molecular dynamics investigations of ligand stabilization via secondary binding site interactions in Paenibacillus xylanivorans GH11 xylanase.
Comput Struct Biotechnol J, 19, 2021
|
|
4URK
 
 | | PI3Kg in complex with AZD6482 | | Descriptor: | 2-[[(1R)-1-(7-methyl-2-morpholin-4-yl-4-oxidanylidene-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Giordanetto, F, Barlaam, B, Berglund, S, Edman, K, Karlsson, O, Lindberg, J, Nylander, S, Inghardt, T. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of 9-(1-Phenoxyethyl)-2-Morpholino-4-Oxo-Pyrido[1, 2-A]Pyrimidine-7-Carboxamides as Oral Pi3Kbeta Inhibitors, Useful as Antiplatelet Agents.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7KZG
 
 | | Human MBD4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
4V96
 
 | | The structure of a 1.8 MDa viral genome injection device suggests alternative infection mechanisms | | Descriptor: | BPP, ORF46, ORF48 | | Authors: | Veesler, D, Spinelli, S, Mahony, J, Lichiere, J, Blangy, S, Bricogne, G, Legrand, P, Ortiz-Lombardia, M, Campanacci, V, van Sinderen, D, Cambillau, C. | | Deposit date: | 2012-02-01 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4WEE
 
 | |
8Q22
 
 | | Crystal structure of Vanadium-dependent haloperoxidase R425S mutant in complex with 1,3,5-trimethoxybenzene (A. marina) | | Descriptor: | 1,3,5-trimethoxybenzene, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Zeides, P, Bellmannn-Sickert, K, Zhang, R, Seel, C.J, Most, V, Schroeder, C.T, Groll, M, Gulder, T. | | Deposit date: | 2023-08-01 | | Release date: | 2024-08-14 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Unraveling the molecular basis of substrate specificity and halogen activation in vanadium-dependent haloperoxidases.
Nat Commun, 16, 2025
|
|
1JB7
 
 | |
1JAK
 
 | | Streptomyces plicatus beta-N-acetylhexosaminidase in Complex with (2R,3R,4S,5R)-2-acetamido-3,4-dihydroxy-5-hydroxymethyl-piperidinium chloride (IFG) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Mark, B.L, Vocadlo, D.J, Zhao, D, Knapp, S, Withers, S.G, James, M.N. | | Deposit date: | 2001-05-30 | | Release date: | 2001-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Biochemical and structural assessment of the 1-N-azasugar GalNAc-isofagomine as a potent family 20 beta-N-acetylhexosaminidase inhibitor.
J.Biol.Chem., 276, 2001
|
|
6P24
 
 | | Escherichia coli tRNA synthetase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-20 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
