7DQT
 
 | | Crystal structure of O6-methylguanine methyltransferase Y91F variant | | Descriptor: | Methylated-DNA--protein-cysteine methyltransferase, SULFATE ION | | Authors: | Kikuchi, M, Iizuka, Y, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2020-12-24 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Roles of the hydroxy group of tyrosine in crystal structures of Sulfurisphaera tokodaii O6-methylguanine-DNA methyltransferase.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7MVS
 
 | | DNA gyrase complexed with uncleaved DNA and Compound 7 to 2.6A resolution | | Descriptor: | (1S)-2-[(2r,5S)-5-{[(2,3-dihydro-1,4-benzodioxin-6-yl)methyl]amino}-1,3-dioxan-2-yl]-1-(3-fluoro-6-methoxyquinolin-4-yl)ethan-1-ol, CHLORIDE ION, DNA (5'-D(P*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Ratigan, S.C, McElroy, C.A. | | Deposit date: | 2021-05-15 | | Release date: | 2021-10-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.60137153 Å) | | Cite: | Optimization of TopoIV Potency, ADMET Properties, and hERG Inhibition of 5-Amino-1,3-dioxane-Linked Novel Bacterial Topoisomerase Inhibitors: Identification of a Lead with In Vivo Efficacy against MRSA.
J.Med.Chem., 64, 2021
|
|
1D53
 
 | | CRYSTAL STRUCTURE AT 1.5 ANGSTROMS RESOLUTION OF D(CGCICICG), AN OCTANUCLEOTIDE CONTAINING INOSINE, AND ITS COMPARISON WITH D(CGCG) AND D(CGCGCG) STRUCTURES | | Descriptor: | DNA (5'-D(*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*IP*CP*IP*CP*G)-3') | | Authors: | Kumar, V.D, Harrison, R.W, Andrews, L.C, Weber, I.T. | | Deposit date: | 1992-11-05 | | Release date: | 1993-04-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure at 1.5-A resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures.
Biochemistry, 31, 1992
|
|
1EH7
 
 | |
1EH6
 
 | | HUMAN O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE | | Descriptor: | O6-ALKYLGUANINE-DNA ALKYLTRANSFERASE, ZINC ION | | Authors: | Daniels, D.S, Tainer, J.A. | | Deposit date: | 2000-02-18 | | Release date: | 2000-04-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active and alkylated human AGT structures: a novel zinc site, inhibitor and extrahelical base binding.
EMBO J., 19, 2000
|
|
7KZG
 
 | | Human MBD4 glycosylase domain bound to DNA containing oxacarbenium-ion analog 1-aza-2'-deoxyribose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pidugu, L.S, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
7KZ1
 
 | | Human MBD4 glycosylase domain bound to DNA containing an abasic site | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*CP*AP*GP*CP*GP*(ORP)P*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*GP*CP*GP*CP*TP*GP*G)-3'), ... | | Authors: | Pidugu, L.S, Bright, H, Pozharski, E, Drohat, A.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insights into the Mechanism of Base Excision by MBD4.
J.Mol.Biol., 433, 2021
|
|
8JNE
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome without the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.68 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8JND
 
 | | The cryo-EM structure of the nonameric RAD51 ring bound to the nucleosome with the linker DNA binding | | Descriptor: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | Authors: | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
4FPV
 
 | | Crystal structure of D. rerio TDP2 complexed with single strand DNA product | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*G)-3'), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, K, Kurahashi, K, Aihara, H. | | Deposit date: | 2012-06-22 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural basis for recognition of 5'-phosphotyrosine adducts by Tdp2.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1EH8
 
 | |
8HYL
 
 | | Crystal structure of DO1 Fv-clasp fragment | | Descriptor: | VH-SARAH, VL-SARAH | | Authors: | Anan, Y, Lu, P, Nagata, K, Itakura, M, Uchida, K. | | Deposit date: | 2023-01-06 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular and structural basis of anti-DNA antibody specificity for pyrrolated proteins.
Commun Biol, 7, 2024
|
|
8R0S
 
 | | Structure of reverse transcriptase from Cauliflower Mosaic Virus in complex with RNA/DNA hybrid | | Descriptor: | DNA (5'-D(*GP*CP*TP*AP*CP*GP*CP*AP*CP*TP*GP*CP*TP*GP*GP*A)-3'), Enzymatic polyprotein, RNA (5'-R(*GP*UP*CP*CP*AP*GP*CP*AP*GP*UP*GP*CP*GP*UP*AP*GP*C)-3') | | Authors: | Prabaharan, C, Figiel, M, Chamera, S, Szczepanowski, R, Nowak, E, Nowotny, M. | | Deposit date: | 2023-10-31 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and biochemical characterization of cauliflower mosaic virus reverse transcriptase.
J.Biol.Chem., 300, 2024
|
|
1DXN
 
 | |
5WW8
 
 | |
5Z51
 
 | |
1ERI
 
 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
5WW4
 
 | |
5WW3
 
 | |
5WW9
 
 | |
8U0U
 
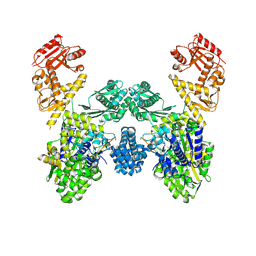 | | DdmD dimer in complex with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*AP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Helicase/UvrB N-terminal domain-containing protein | | Authors: | Bravo, J.P.K. | | Deposit date: | 2023-08-29 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Plasmid targeting and destruction by the DdmDE bacterial defence system.
Nature, 630, 2024
|
|
8IA3
 
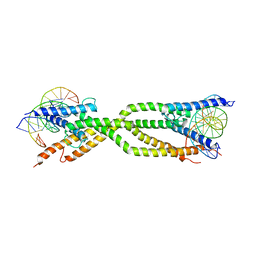 | | Crystal structure of human USF2 bHLHLZ domain in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*TP*CP*AP*CP*GP*TP*GP*CP*CP*CP*GP*TP*C)-3'), DNA (5'-D(P*GP*AP*CP*GP*GP*GP*CP*AP*CP*GP*TP*GP*AP*CP*GP*CP*GP*C)-3'), Upstream stimulatory factor 2 | | Authors: | Huang, C, Fang, P, Wang, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Tetramerization of upstream stimulating factor USF2 requires the elongated bent leucine zipper of the bHLH-LZ domain.
J.Biol.Chem., 299, 2023
|
|
1D9F
 
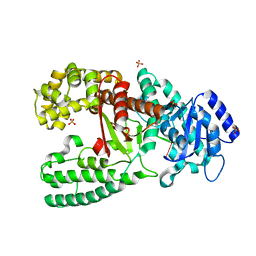 | | CRYSTAL STRUCTURE OF THE COMPLEX OF DNA POLYMERASE I KLENOW FRAGMENT WITH DNA TETRAMER CARRYING 2'-O-(3-AMINOPROPYL)-RNA MODIFICATION 5'-D(TT)-AP(U)-D(T)-3' | | Descriptor: | DNA POLYMERASE I, DNA/RNA (5'-D(*TP*TP)-R(*(U31)P)-D(*T)-3'), SULFATE ION, ... | | Authors: | Teplova, M, Wallace, S.T, Tereshko, V, Minasov, G, Simons, A.M, Cook, P.D, Manoharan, M, Egli, M. | | Deposit date: | 1999-10-27 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural origins of the exonuclease resistance of a zwitterionic RNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5WW7
 
 | |
5WW5
 
 | |
