1HHU
 
 | | Balhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, BALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertessy, L. | | Deposit date: | 2000-12-28 | | Release date: | 2003-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1HHV
 
 | | SOLUTION STRUCTURE OF VIRUS CHEMOKINE VMIP-II | | Descriptor: | VIRUS CHEMOKINE VMIP-II | | Authors: | Shao, W, Fernandez, E, Navenot, J.M, Wilken, J, Thompson, D.A, Pepiper, S, Schweitzer, B.I, Lolis, E. | | Deposit date: | 1998-12-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | CCR2 and CCR5 receptor-binding properties of herpesvirus-8
vMIP-II based on sequence analysis and its solution structure
Eur.J.Biochem., 268, 2001
|
|
1HHW
 
 | | Solution structure of LNA1:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1HHX
 
 | | Solution structure of LNA3:RNA hybrid | | Descriptor: | 5- D(*CP*+TP*GP*AP*+TP*AP*+TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1HHY
 
 | | Deglucobalhimycin in complex with D-Ala-D-Ala | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, D-ALANINE, DEGLUCOBALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertesy, L. | | Deposit date: | 2000-12-29 | | Release date: | 2003-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1HHZ
 
 | | Deglucobalhimycin in complex with cell wall pentapeptide | | Descriptor: | (2R,4S,6S)-4-azanyl-4,6-dimethyl-oxane-2,5,5-triol, CELL WALL PEPTIDE, DEGLUCOBALHIMYCIN, ... | | Authors: | Lehmann, C, Bunkoczi, G, Sheldrick, G.M, Vertesy, L. | | Deposit date: | 2000-12-29 | | Release date: | 2003-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structures of Glycopeptide Antibiotics with Peptides that Model Bacterial Cell-Wall Precursors
J.Mol.Biol., 318, 2002
|
|
1HI0
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus initiation complex | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-31 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HI1
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus bound NTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-01 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HI2
 
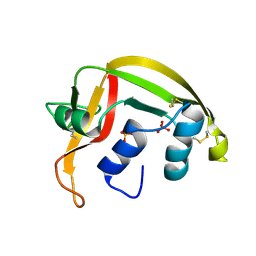 | | Eosinophil-derived Neurotoxin (EDN) - Sulphate Complex | | Descriptor: | EOSINOPHIL-DERIVED NEUROTOXIN, SULFATE ION | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI3
 
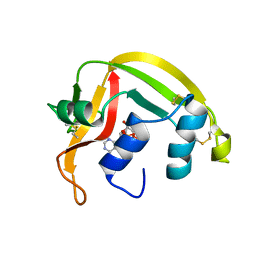 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine 2'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI4
 
 | | Eosinophil-derived Neurotoxin (EDN) - Adenosien-3'-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI5
 
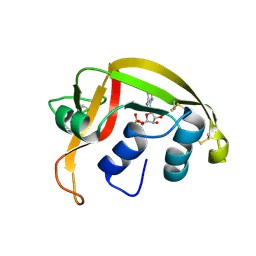 | | Eosinophil-derived Neurotoxin (EDN) - Adenosine-5'-Diphosphate Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Leonidas, D.D, Boix, E, Prill, R, Suzuki, M, Turton, R, Minson, K, Swaminathan, G.J, Youle, R.J, Acharya, K.R. | | Deposit date: | 2001-01-02 | | Release date: | 2001-05-31 | | Last modified: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mapping the Ribonucleolytic Active Site of Eosinophil-Derived Neurotoxin (Edn): High Resolution Crystal Structures of Edn Complexes with Adenylic Nucleotide Inhibitors
J.Biol.Chem., 276, 2001
|
|
1HI6
 
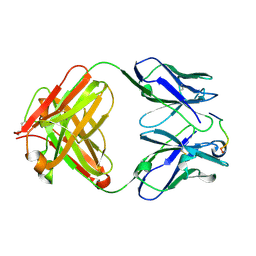 | | ANTI-P24 (HIV-1) FAB FRAGMENT CB41 COMPLEXED WITH A PEPTIDE | | Descriptor: | IGG2A KAPPA ANTIBODY CB41 (HEAVY CHAIN), IGG2A KAPPA ANTIBODY CB41 (LIGHT CHAIN), PEPTIDE 5 | | Authors: | Hahn, M, Wessner, H, Schneider-Mergener, J, Hohne, W. | | Deposit date: | 2001-01-02 | | Release date: | 2001-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic Analysis of Anti-P24 (HIV-1) Monoclonal Antibody Cross-Reactivity and Polyspecificity
Cell(Cambridge,Mass.), 91, 1997
|
|
1HI7
 
 | |
1HI8
 
 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MAGNESIUM ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-27 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HI9
 
 | | Zn-dependent D-aminopeptidase DppA from Bacillus subtilis, a self-compartmentalizing protease. | | Descriptor: | DIPEPTIDE TRANSPORT PROTEIN DPPA, ZINC ION | | Authors: | Remaut, H, Bompard-Gilles, C, Goffin, C, Frere, J.M, Van Beeumen, J. | | Deposit date: | 2001-01-04 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Bacillus Subtilis D-Aminopeptidase Dppa Reveals a Novel Self-Compartmentalizing Protease
Nat.Struct.Biol., 8, 2001
|
|
1HIA
 
 | | KALLIKREIN COMPLEXED WITH HIRUSTASIN | | Descriptor: | HIRUSTASIN, KALLIKREIN | | Authors: | Mittl, P, Di Marco, S, Gruetter, M. | | Deposit date: | 1996-12-12 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A new structural class of serine protease inhibitors revealed by the structure of the hirustasin-kallikrein complex.
Structure, 5, 1997
|
|
1HIB
 
 | | THE STRUCTURE OF AN INTERLEUKIN-1 BETA MUTANT WITH REDUCED BIOACTIVITY SHOWS MULTIPLE SUBTLE CHANGES IN CONFORMATION THAT AFFECT PROTEIN-PROTEIN RECOGNITION | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Camacho, N.P, Smith, D.R, Goldman, A, Schneider, B, Green, D, Young, P.R, Berman, H.M. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an interleukin-1 beta mutant with reduced bioactivity shows multiple subtle changes in conformation that affect protein-protein recognition.
Biochemistry, 32, 1993
|
|
1HIC
 
 | |
1HIG
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN INTERFERON-GAMMA. | | Descriptor: | INTERFERON-GAMMA | | Authors: | Ealick, S.E, Cook, W.J, Vijay-Kumar, S, Carson, M, Nagabhushan, T.L, Trotta, P.P, Bugg, C.E. | | Deposit date: | 1991-10-03 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Three-dimensional structure of recombinant human interferon-gamma.
Science, 252, 1991
|
|
1HIH
 
 | |
1HII
 
 | |
1HIJ
 
 | |
1HIK
 
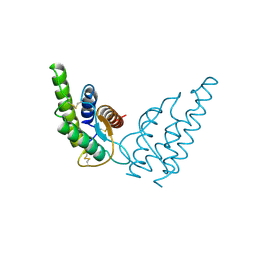 | | INTERLEUKIN-4 (WILD-TYPE) | | Descriptor: | INTERLEUKIN-4 | | Authors: | Mueller, T, Buehner, M. | | Deposit date: | 1995-06-01 | | Release date: | 1996-01-29 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human interleukin-4 and variant R88Q: phasing X-ray diffraction data by molecular replacement using X-ray and nuclear magnetic resonance models.
J.Mol.Biol., 247, 1995
|
|
1HIL
 
 | |
