3NL1
 
 | | Crystal Structure of Salicylate 1,2-dioxygenase from Pseudoaminobacter salicylatoxidans Adducts with gentisate | | Descriptor: | 2,5-dihydroxybenzoic acid, FE (II) ION, GLYCEROL, ... | | Authors: | Ferraroni, M, Briganti, F, Matera, I. | | Deposit date: | 2010-06-21 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Crystal structures of salicylate 1,2-dioxygenase-substrates adducts: A step towards the comprehension of the structural basis for substrate selection in class III ring cleaving dioxygenases.
J.Struct.Biol., 177, 2012
|
|
3NEA
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Francisella tularensis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Lam, R, McGrath, T.E, Romanov, V, Gothe, S.A, Peddi, S.R, Razumova, E, Lipman, R.S, Branstrom, A.A, Chirgadze, N.Y. | | Deposit date: | 2010-06-08 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Francisella tularensis peptidyl-tRNA hydrolase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3NHH
 
 | | Crystal structure of Staphylococcal nuclease variant V23E-L36K at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Robinson, A.C, Schlessman, J.L, Garcia-Moreno, E.B. | | Deposit date: | 2010-06-14 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic consequences of burial of an artificial ion pair in the hydrophobic interior of a protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3NHX
 
 | | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound | | Descriptor: | 4-ANDROSTENE-3-17-DIONE, SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Ruben, E, Sunden, F, Herschlag, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D99N from Pseudomonas Testosteroni (tKSI) with 4-Androstene-3,17-dione Bound
To be Published
|
|
3NI3
 
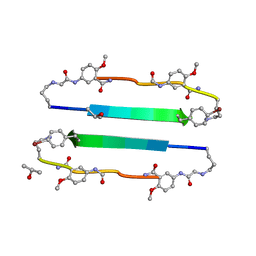 | | 54-Membered ring macrocyclic beta-sheet peptide | | Descriptor: | 54-membered ring macrocyclic beta-sheet peptide, ISOPROPYL ALCOHOL | | Authors: | Sawaya, M.R, Eisenberg, D, Nowick, J.S, Korman, T.P, Khakshoor, O. | | Deposit date: | 2010-06-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | X-ray crystallographic structure of an artificial beta-sheet dimer.
J.Am.Chem.Soc., 132, 2010
|
|
3N8B
 
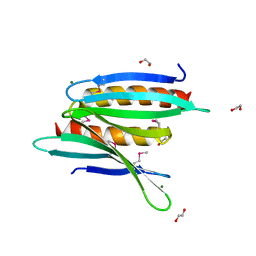 | | Crystal Structure of Borrelia burgdorferi Pur-alpha | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Graebsch, A, Roche, S, Kostrewa, D, Niessing, D. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-06 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Of bits and bugs--on the use of bioinformatics and a bacterial crystal structure to solve a eukaryotic repeat-protein structure.
Plos One, 5, 2010
|
|
3NEO
 
 | |
3NEY
 
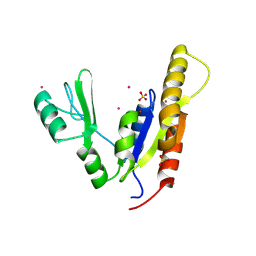 | | Crystal structure of the kinase domain of MPP1/p55 | | Descriptor: | 55 kDa erythrocyte membrane protein, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tong, Y, Zhong, N, Guan, X, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the kinase domain of MPP1/p55
To be Published
|
|
3NFY
 
 | |
3NEV
 
 | | Crystal structure of YagE, a prophage protein from E. coli K12 in complex with KDGal | | Descriptor: | 1,2-ETHANEDIOL, 3-DEOXY-D-LYXO-HEXONIC ACID, Uncharacterized protein yagE | | Authors: | Bhaskar, V, Kumar, P.M, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of biochemical and putative biological role of a xenolog from Escherichia coli using structural analysis.
Proteins, 79, 2011
|
|
3NFZ
 
 | | Crystal structure of murine aminoacylase 3 in complex with N-acetyl-L-tyrosine | | Descriptor: | Aspartoacylase-2, CHLORIDE ION, N-acetyl-L-tyrosine, ... | | Authors: | Hsieh, J.M, Tsirulnikov, K, Sawaya, M.R, Magilnick, N, Abuladze, N, Kurtz, I, Abramson, J, Pushkin, A. | | Deposit date: | 2010-06-10 | | Release date: | 2010-10-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structures of aminoacylase 3 in complex with acetylated substrates.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NAQ
 
 | | Apo-form of NAD-dependent formate dehydrogenase from higher-plant Arabidopsis thaliana | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Shabalin, I.G, Polyakov, K.M, Serov, A.E, Skirgello, O.E, Sadykhov, E.G, Dorovatovskiy, P.V, Tishkov, V.I, Popov, V.O. | | Deposit date: | 2010-06-02 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the apo and holo forms of NAD-dependent formate dehydrogenase from the higher-plant Arabidopsis Thaliana
to be published
|
|
3NA5
 
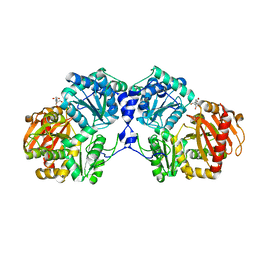 | | Crystal structure of a bacterial phosphoglucomutase, an enzyme important in the virulence of several human pathogens. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, Phosphoglucomutase | | Authors: | Mehra-Chaudhary, R, Beamer, L.J. | | Deposit date: | 2010-06-01 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens.
Proteins, 79, 2011
|
|
3NFW
 
 | |
3NCC
 
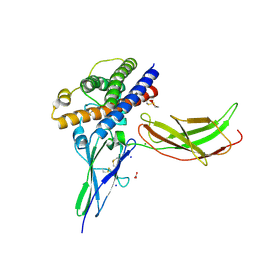 | | A human Prolactin receptor antagonist in complex with the mutant extracellular domain H188A of the human prolactin receptor | | Descriptor: | CARBONATE ION, CHLORIDE ION, Prolactin, ... | | Authors: | Kulkarni, M.V, Tettamanzi, M.C, Murphy, J.W, Keeler, C, Myszka, D.G, Chayen, N.E, Lolis, E.J, Hodsdon, M.E. | | Deposit date: | 2010-06-04 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two Independent Histidines, One in Human Prolactin and One in Its Receptor, Are Critical for pH-dependent Receptor Recognition and Activation.
J.Biol.Chem., 285, 2010
|
|
3ND0
 
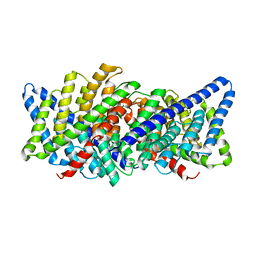 | | X-ray crystal structure of a slow cyanobacterial Cl-/H+ antiporter | | Descriptor: | CHLORIDE ION, Sll0855 protein | | Authors: | Jayaram, H, Robertson, J, Wu, F, Williams, C, Miller, C. | | Deposit date: | 2010-06-06 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a Slow CLC Cl(-)/H(+) Antiporter from a Cyanobacterium.
Biochemistry, 50, 2011
|
|
3NDN
 
 | |
3NJ6
 
 | | 0.95 A resolution X-ray structure of (GGCAGCAGCC)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*GP*CP*AP*GP*CP*C)-3', SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Atomic resolution structure of CAG RNA repeats: structural insights and implications for the trinucleotide repeat expansion diseases.
Nucleic Acids Res., 38, 2010
|
|
3NJF
 
 | | Y26F mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJY
 
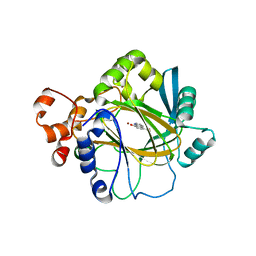 | | Crystal structure of JMJD2A complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 8-hydroxyquinoline-5-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | King, O.N.F, Clifton, I.J, Wang, M, Maloney, D.J, Jadhav, A, Oppermann, U, Heightman, T.D, Simeonov, A, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2010-06-18 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Quantitative high-throughput screening identifies 8-hydroxyquinolines as cell-active histone demethylase inhibitors
Plos One, 5, 2010
|
|
3NMZ
 
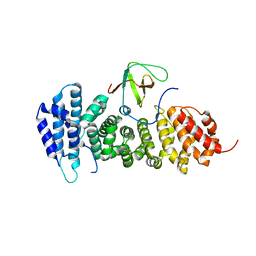 | | Crystal structure of APC complexed with Asef | | Descriptor: | APC variant protein, Rho guanine nucleotide exchange factor 4 | | Authors: | Zhang, Z, Chen, L, Gao, L, Lin, K, Wu, G. | | Deposit date: | 2010-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural basis for the recognition of Asef by adenomatous polyposis coli.
Cell Res., 22, 2012
|
|
3N9G
 
 | |
3NBQ
 
 | |
3NC1
 
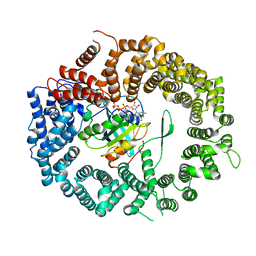 | | Crystal structure of the CRM1-RanGTP complex | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Guttler, T, Madl, T, Neumann, P, Deichsel, D, Corsini, L, Monecke, T, Ficner, R, Sattler, M, Gorlich, D. | | Deposit date: | 2010-06-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1.
Nat.Struct.Mol.Biol., 17, 2010
|
|
