6L8E
 
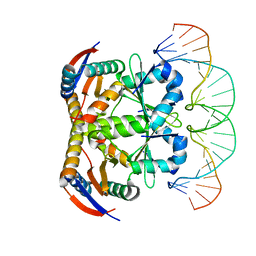 | | Crystal structure of heterohexameric YoeB-YefM complex bound to 26bp-DNA | | Descriptor: | DNA (26-mer), YefM Antitoxin, YoeB toxin | | Authors: | Yue, J, Xue, L. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Distinct oligomeric structures of the YoeB-YefM complex provide insights into the conditional cooperativity of type II toxin-antitoxin system.
Nucleic Acids Res., 48, 2020
|
|
6WMT
 
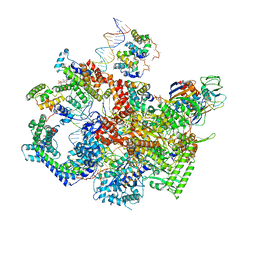 | | F. tularensis RNAPs70-(MglA-SspA)-ppGpp-PigR-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA NT-strand downstream, DNA T-strand, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
6BZO
 
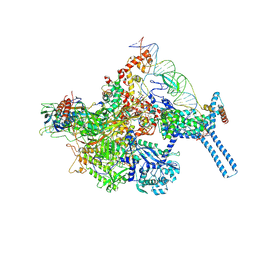 | | Mtb RNAP Holo/RbpA/Fidaxomicin/upstream fork DNA | | Descriptor: | DNA (26-MER), DNA (32-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J. | | Deposit date: | 2017-12-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
6C04
 
 | | Mtb RNAP Holo/RbpA/double fork DNA -closed clamp | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Darst, S.A, Campbell, E.A, Boyaci Selcuk, H, Chen, J, Lilic, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Fidaxomicin jamsMycobacterium tuberculosisRNA polymerase motions needed for initiation via RbpA contacts.
Elife, 7, 2018
|
|
7W5Y
 
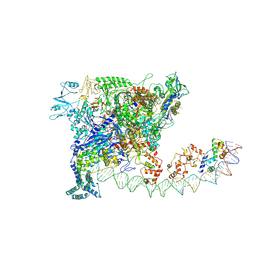 | |
7W5W
 
 | |
5B2I
 
 | | Human nucleosome containing CpG unmethylated DNA | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Fujii, Y, Wakamori, M, Umehara, T, Yokoyama, S. | | Deposit date: | 2016-01-16 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human nucleosome core particle containing enzymatically introduced CpG methylation.
Febs Open Bio, 6, 2016
|
|
5XFQ
 
 | | Ternary complex of PHF1, a DNA duplex and a histone peptide | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*GP*CP*CP*GP*CP*CP*CP*T)-3'), PHD finger protein 1, Peptide from Histone H3, ... | | Authors: | Wang, Z, Li, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Polycomb-like proteins link the PRC2 complex to CpG islands
Nature, 549, 2017
|
|
7W5X
 
 | | Cryo-EM structure of SoxS-dependent transcription activation complex with zwf promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of three different transcription activation strategies adopted by a single regulator SoxS.
Nucleic Acids Res., 50, 2022
|
|
2DAU
 
 | | DICKERSON-DREW DNA DODECAMER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Denisov, A, Bekiroglu, S, Maltseva, T, Sandstrom, A, Altmann, K.-H, Egli, M, Chattopadhyaya, J. | | Deposit date: | 1998-01-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution conformation of a carbocyclic analog of the Dickerson-Drew dodecamer: comparison with its own X-ray structure and that of the NMR structure of the native counterpart.
J.Biomol.Struct.Dyn., 16, 1998
|
|
6J50
 
 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome (tilted conformation) | | Descriptor: | DNA (198-MER), DNA (41-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J4Z
 
 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
6J51
 
 | | RNA polymerase II elongation complex bound with Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome, weak Elf1 (+1 position) | | Descriptor: | DNA (198-MER), DNA (36-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Ehara, H, Kujirai, T, Fujino, Y, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science, 363, 2019
|
|
1EA4
 
 | | TRANSCRIPTIONAL REPRESSOR COPG/22bp dsDNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*CP*GP*TP*GP *CP*AP*CP*TP*CP*AP*AP*TP*GP*CP*AP*AP*TP*C)-3'), DNA(5'-D(*AP*GP*AP*TP*TP*GP*CP*AP*TP *TP*GP*AP*GP*TP*GP*CP*AP*CP*GP*GP*TP*T)-3'), TRANSCRIPTIONAL REPRESSOR COPG | | Authors: | Gomis-Rueth, F.X, Costa, M, Sola, M, Acebo, P, Eritja, R, Espinosa, M, Solar, G.D, Coll, M. | | Deposit date: | 2000-11-05 | | Release date: | 2001-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plasmid Transcriptional Repressor Copg Oligomerises to Render Helical Superstructures Unbound and in Complexes with Oligonucleotides
J.Mol.Biol., 310, 2001
|
|
6INQ
 
 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA (+1 position) | | Descriptor: | DNA (198-MER), DNA (31-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-10-26 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
7UM0
 
 | | Structure of the phage AR9 non-virion RNA polymerase holoenzyme in complex with two DNA oligonucleotides containing the AR9 P077 promoter as determined by cryo-EM | | Descriptor: | DNA (5'-D(P*GP*UP*U)-3'), DNA-directed RNA polymerase, DNA-directed RNA polymerase beta subunit, ... | | Authors: | Leiman, P.G, Fraser, A, Sokolova, M.L. | | Deposit date: | 2022-04-05 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of template strand deoxyuridine promoter recognition by a viral RNA polymerase.
Nat Commun, 13, 2022
|
|
6WMP
 
 | | F. tularensis RNAPs70-iglA DNA complex | | Descriptor: | DNA NT-strand, DNA T-strand, DNA-directed RNA polymerase subunit alpha 1, ... | | Authors: | Travis, B.A, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural Basis for Virulence Activation of Francisella tularensis.
Mol.Cell, 81, 2021
|
|
7F0R
 
 | | Cryo-EM structure of Pseudomonas aeruginosa SutA transcription activation complex | | Descriptor: | DNA (70-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-06-06 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
4WZW
 
 | | Crystal structure of human Puf-A in complex with DNA | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), Pumilio domain-containing protein KIAA0020 | | Authors: | Qiu, C, Hall, T.M.T. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9516 Å) | | Cite: | A divergent Pumilio repeat protein family for pre-rRNA processing and mRNA localization.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
9GS9
 
 | | Tn7016 PseCAST QCascade | | Descriptor: | Cas6, Cas7.1, Cas8, ... | | Authors: | Lampe, G.D, Liang, A.R, Zhang, D.J, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided engineering of type I-F CASTs for targeted gene insertion in human cells.
Biorxiv, 2024
|
|
6EU1
 
 | | RNA Polymerase III - open DNA complex (OC-POL3). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
8YBJ
 
 | | Cryo-EM structure of human nucleosome core particle composed of the Widom 601 DNA sequence | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kimura, T, Hirai, S, Kujirai, T, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and biochemical analyses of the nucleosome containing the cancer-associated histone H3 mutation E97K.
Genes Cells, 29, 2024
|
|
