2MDQ
 
 | |
3J8B
 
 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S-HCV IRES EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
3J8C
 
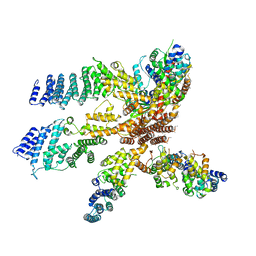 | | Model of the human eIF3 PCI-MPN octamer docked into the 43S EM map | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit C, Eukaryotic translation initiation factor 3 subunit E, ... | | Authors: | Erzberger, J.P, Ban, N. | | Deposit date: | 2014-10-08 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell(Cambridge,Mass.), 158, 2014
|
|
2OSH
 
 | |
2PCO
 
 | | Spatial Structure and Membrane Permeabilization for Latarcin-1, a Spider Antimicrobial Peptide | | Descriptor: | Latarcin-1 | | Authors: | Dubovskii, P.V, Volynsky, P.E, Polyansky, A.A, Chupin, V.V, Efremov, R.G, Arseniev, A.S. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure/hydrophobicity of latarcins specifies their mode of membrane activity.
Biochemistry, 47, 2008
|
|
2OWO
 
 | | Last Stop on the Road to Repair: Structure of E.coli DNA Ligase Bound to Nicked DNA-Adenylate | | Descriptor: | 26-MER, 5'-D(*AP*CP*AP*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*G)-3', ... | | Authors: | Shuman, S, Nandakumar, J, Nair, P.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Last Stop on the Road to Repair: Structure of E. coli DNA Ligase Bound to Nicked DNA-Adenylate.
Mol.Cell, 26, 2007
|
|
2LKB
 
 | | Evolutionary diversification of Mesobuthus alpha-scorpion toxins affecting sodium channels | | Descriptor: | Neurotoxin MeuNaTx-5 | | Authors: | Zhu, S, Peigneur, S, Gao, B, Lu, X, Cao, C, Tytgat, J. | | Deposit date: | 2011-10-09 | | Release date: | 2011-11-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Evolutionary diversification of Mesobuthus {alpha}-scorpion toxins affecting sodium channels
Mol.Cell Proteomics, 2011
|
|
7EP8
 
 | |
7O0N
 
 | |
3OKG
 
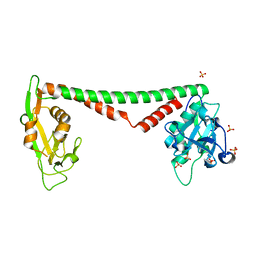 | | Crystal structure of HsdS subunit from Thermoanaerobacter tengcongensis | | Descriptor: | Restriction endonuclease S subunits, SULFATE ION | | Authors: | Liang, D, Gao, P, Tang, Q, An, X, Yan, X. | | Deposit date: | 2010-08-24 | | Release date: | 2011-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of HsdS subunit from Thermoanaerobacter tengcongensis sheds lights on mechanism of dynamic opening and closing of type I methyltransferase
Plos One, 6, 2011
|
|
3OT0
 
 | |
7K33
 
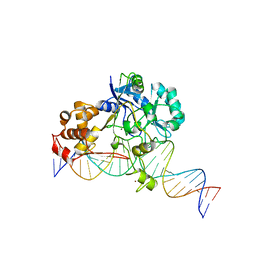 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
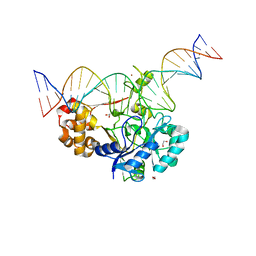 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | Descriptor: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K32
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | Descriptor: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | Authors: | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | Deposit date: | 2020-09-10 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KD9
 
 | | Crystal Structure of Gallic Acid Decarboxylase from Arxula adeninivorans | | Descriptor: | Gallate decarboxylase, POTASSIUM ION | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMR
 
 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pea pathogenicity protein 2 | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-02-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
7JMV
 
 | | Crystal structure of the pea pathogenicity protein 2 from Madurella mycetomatis complexed with 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Zeug, M, Markovic, N, Iancu, C.V, Tripp, J, Oreb, M, Choe, J. | | Deposit date: | 2020-08-03 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of non-oxidative decarboxylases reveal a new mechanism of action with a catalytic dyad and structural twists.
Sci Rep, 11, 2021
|
|
3SNZ
 
 | |
1CZ1
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1C52
 
 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
1EQC
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS IN COMPLEX WITH CASTANOSPERMINE AT 1.85 A | | Descriptor: | CASTANOSPERMINE, EXO-(B)-(1,3)-GLUCANASE | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
1FJE
 
 | | SOLUTION STRUCTURE OF NUCLEOLIN RBD12 IN COMPLEX WITH SNRE RNA | | Descriptor: | NUCLEOLIN RBD12, SNRE RNA | | Authors: | Allain, F.H.T, Bouvet, P, Dieckmann, T, Feigon, J. | | Deposit date: | 2000-08-08 | | Release date: | 2001-01-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin.
EMBO J., 19, 2000
|
|
1GID
 
 | | CRYSTAL STRUCTURE OF A GROUP I RIBOZYME DOMAIN: PRINCIPLES OF RNA PACKING | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, P4-P6 RNA RIBOZYME DOMAIN | | Authors: | Cate, J.H, Gooding, A.R, Podell, E, Zhou, K, Golden, B.L, Kundrot, C.E, Cech, T.R, Doudna, J.A. | | Deposit date: | 1996-08-22 | | Release date: | 1996-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a group I ribozyme domain: principles of RNA packing.
Science, 273, 1996
|
|
