6L15
 
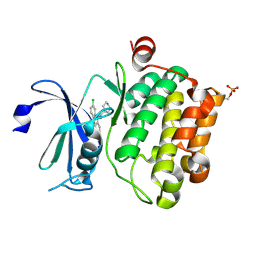 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
3RLZ
 
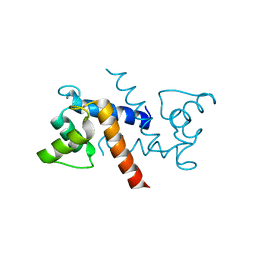 | |
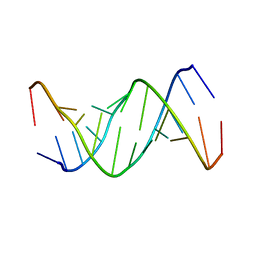
1ELH
 
 | |
1EWW
 
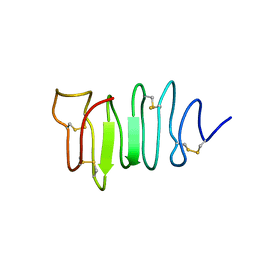 | | SOLUTION STRUCTURE OF SPRUCE BUDWORM ANTIFREEZE PROTEIN AT 30 DEGREES CELSIUS | | Descriptor: | ANTIFREEZE PROTEIN | | Authors: | Graether, S.P, Kuiper, M.J, Gagne, S.M, Walker, V.K, Jia, Z, Sykes, B.D, Davies, P.L. | | Deposit date: | 2000-04-27 | | Release date: | 2000-07-27 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Beta-helix structure and ice-binding properties of a hyperactive antifreeze protein from an insect.
Nature, 406, 2000
|
|
3RXB
 
 | | Crystal structure of Trypsin complexed with 4-guanidinobutanoic acid | | Descriptor: | 4-carbamimidamidobutanoic acid, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXP
 
 | | Crystal structure of Trypsin complexed with (1,5-dimethylpyrazol-3-yl)methanamine | | Descriptor: | 1-(1,5-dimethyl-1H-pyrazol-3-yl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
3RXD
 
 | | Crystal structure of Trypsin complexed with (3-methoxyphenyl)methanamine | | Descriptor: | 1-(3-methoxyphenyl)methanamine, CALCIUM ION, Cationic trypsin, ... | | Authors: | Yamane, J, Yao, M, Zhou, Y, Tanaka, I. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In-crystal affinity ranking of fragment hit compounds reveals a relationship with their inhibitory activities
J.Appl.Crystallogr., 44, 2011
|
|
6L2H
 
 | | CGTase mutant-Y167H | | Descriptor: | Alpha-cyclodextrin glucanotransferase, CALCIUM ION | | Authors: | Fan, T.W, Hou, A.Q, Chao, Y.P, Sun, Y. | | Deposit date: | 2019-10-03 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structure basis of a mutant a-CGTase tyrosine167histidine from Bacillus sp. 602-1 with enhanced a-CD production
To Be Published
|
|
1EZ3
 
 | |
3PMV
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Jamieson, C, Brown, C.I, Campbell, R.A, Gillen, K.J, Gillespie, J, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure based evolution of a novel series of positive modulators of the AMPA receptor.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6XMA
 
 | | Crystal structure of iron-bound LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | Dioxygenase, FE (III) ION, SULFATE ION | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6XM9
 
 | | Crystal structure of vanillin bound to Co-LSD4 from Sphingobium sp. strain SYK-6 | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, ACETATE ION, COBALT (II) ION, ... | | Authors: | Kuatsjah, E, Chan, A.C, Katahira, R, Beckham, G.T, Murphy, M.E, Eltis, L.D. | | Deposit date: | 2020-06-29 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and functional analysis of lignostilbene dioxygenases from Sphingobium sp. SYK-6.
J.Biol.Chem., 296, 2021
|
|
6L3W
 
 | | Crystal structure of BphC, a halotolerant catechol dioxygenase | | Descriptor: | Extra-diol dioxygenase BphC, FE (III) ION | | Authors: | Thakur, K.G, Solanki, V. | | Deposit date: | 2019-10-15 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure Elucidation and Biochemical Characterization of Environmentally Relevant Novel Extradiol Dioxygenases Discovered by a Functional Metagenomics Approach.
mSystems, 4, 2019
|
|
6L1R
 
 | |
3QT3
 
 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Clostridium perfringens CPE0426 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein CPE0426 | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-22 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
3QVS
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus wild type | | Descriptor: | GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
6XI5
 
 | |
6XI4
 
 | |
3QW2
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus mutant N255A | | Descriptor: | GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-26 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
6L5B
 
 | | The structure of the UdgX mutant H109E at a post-excision state | | Descriptor: | IRON/SULFUR CLUSTER, Uracil DNA glycosylase superfamily protein | | Authors: | Xie, W, Tu, J, Zeng, H. | | Deposit date: | 2019-10-22 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00004983 Å) | | Cite: | Structural insights into an MsmUdgX mutant capable of both crosslinking and uracil excision capability.
DNA Repair (Amst), 97, 2021
|
|
2HL9
 
 | |
6XNE
 
 | |
6A35
 
 | |
6XNM
 
 | |
