1Y9H
 
 | | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*CP*CP*AP*TP*(5CM)P*(BPG)P*CP*TP*AP*CP*C)-3', 5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3' | | Authors: | Zhang, N, Lin, C, Huang, X, Kolbanovskiy, A, Hingerty, B.E, Amin, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-22 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-N2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement.
J.Mol.Biol., 346, 2005
|
|
3QF7
 
 | |
1K2L
 
 | | STRUCTURAL CHARACTERIZATION OF BISINTERCALATION IN HIGHER-ORDER DNA AT A JUNCTION-LIKE QUADRUPLEX | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*G)-3', BIS-(9-OCTYLAMINO(2-DIMETHYLAMINOETHYL)ACRIDINE-4-CARBOXAMIDE, COBALT (II) ION | | Authors: | Teixeira, S.C.M, Thorpe, J.H, Todd, A.K, Powell, H.R, Adams, A, Wakelin, L.P.G, Denny, W.A, Cardin, C.J. | | Deposit date: | 2001-09-28 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterisation of Bisintercalation in Higher-order DNA at a Junction-like Quadruplex
J.MOL.BIOL., 323, 2002
|
|
1OLD
 
 | |
4E60
 
 | |
3OOR
 
 | | I-SceI mutant (K86R/G100T)complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
1X51
 
 | | Solution structure of the NUDIX domain from human A/G-specific adenine DNA glycosylase alpha-3 splice isoform | | Descriptor: | A/G-specific adenine DNA glycosylase | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NUDIX domain from human A/G-specific adenine DNA glycosylase alpha-3 splice isoform
To be Published
|
|
1S9O
 
 | | Solution structure of the nitrous acid induced DNA interstrand cross-linked dodecamer duplex CGCTAC(G)TAGCG with the cross-linked guanines denoted (G) | | Descriptor: | 5'-D(*CP*GP*CP*TP*AP*CP*GP*TP*AP*GP*CP*G)-3' | | Authors: | Edfeldt, N.B, Harwood, E.A, Sigurdsson, S.T, Hopkins, P.B, Reid, B.R. | | Deposit date: | 2004-02-05 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a nitrous acid induced DNA interstrand cross-link.
Nucleic Acids Res., 32, 2004
|
|
1XCZ
 
 | | Structure of DNA containing the beta-anomer of a carbocyclic abasic site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(DXD)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, El-khateeb, M, Rege, P, Tian, K, Johnson, F. | | Deposit date: | 2004-09-03 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of the C1 configuration of abasic sites on DNA duplex structure
Biochemistry, 43, 2004
|
|
1XCY
 
 | | Structure of DNA containing the alpha-anomer of a carbocyclic abasic site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(DXD)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, El-khateeb, M, Rege, P, Tian, K, Johnson, F. | | Deposit date: | 2004-09-03 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Impact of the C1 configuration of abasic sites on DNA duplex structure
Biochemistry, 43, 2004
|
|
1DDP
 
 | | Solution structure of a CISPLATIN-INDUCED [CATAGCTATG]2 Interstrand cross-link | | Descriptor: | Cisplatin, DNA (5'-D(*CP*AP*TP*AP*GP*CP*TP*AP*TP*G)-3') | | Authors: | Zhu, L, Huang, H, Reid, B.R, Drobny, G.P, Hopkins, P.B. | | Deposit date: | 1995-10-26 | | Release date: | 1996-03-08 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a cisplatin-induced DNA interstrand cross-link.
Science, 270, 1995
|
|
1SM5
 
 | | Crystal Structure of a DNA Decamer Containing a Thymine-dimer | | Descriptor: | 5'-D(*CP*GP*AP*AP*TP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*(BRU)P*TP*AP*AP*TP*(BRU)P*CP*G)-3' | | Authors: | Park, H, Zhang, K, Ren, Y, Nadji, S, Sinha, N, Taylor, J.-S, Kang, C. | | Deposit date: | 2004-03-08 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a DNA Decamer Containing a Thymine-dimer
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2HMY
 
 | | BINARY COMPLEX OF HHAI METHYLTRANSFERASE WITH ADOMET FORMED IN THE PRESENCE OF A SHORT NONPSECIFIC DNA OLIGONUCLEOTIDE | | Descriptor: | PROTEIN (CYTOSINE-SPECIFIC METHYLTRANSFERASE HHAI), S-ADENOSYLMETHIONINE | | Authors: | O'Gara, M, Zhang, X, Roberts, R.J, Cheng, X. | | Deposit date: | 1999-02-08 | | Release date: | 1999-03-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of a binary complex of HhaI methyltransferase with S-adenosyl-L-methionine formed in the presence of a short non-specific DNA oligonucleotide.
J.Mol.Biol., 287, 1999
|
|
3UYH
 
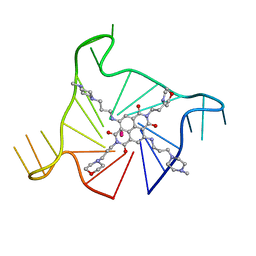 | | Crystal structure of an intramolecular human telomeric DNA G-quadruplex bound by the naphthalene diimide compound, MM41 | | Descriptor: | 4,9-bis{[3-(4-methylpiperazin-1-yl)propyl]amino}-2,7-bis[3-(morpholin-4-yl)propyl]benzo[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetrone, POTASSIUM ION, human telomeric DNA sequence | | Authors: | Collie, G.W, Neidle, S. | | Deposit date: | 2011-12-06 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design and evaluation of naphthalene diimide g-quadruplex ligands as telomere targeting agents in pancreatic cancer cells.
J.Med.Chem., 56, 2013
|
|
1HMY
 
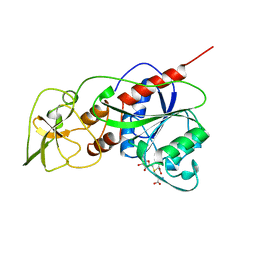 | |
1RES
 
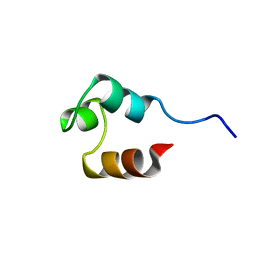 | |
1RET
 
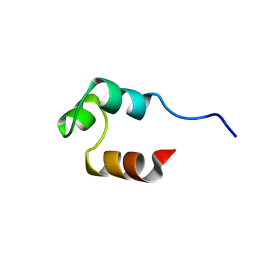 | |
3I6W
 
 | |
2ODH
 
 | | Restriction Endonuclease BCNI in the Absence of DNA | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, R.BcnI | | Authors: | Sokolowska, M, Kaus-Drobek, M, Czapinska, H, Tamulaitis, G, Szczepanowski, R.H, Urbanke, K, Siksnys, V, Bochtler, M. | | Deposit date: | 2006-12-22 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Monomeric restriction endonuclease BcnI in the apo form and in an asymmetric complex with target DNA.
J.Mol.Biol., 369, 2007
|
|
4D9X
 
 | | The crystal structure of Coptisine bound to DNA d(CGTACG) | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The crystal structure of Coptisine bound to DNA d(CGTACG)
to be published
|
|
1JU0
 
 | |
3I6U
 
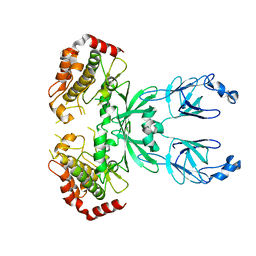 | |
4HYM
 
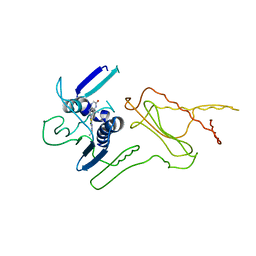 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 7-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)pyrido[2,3-b]pyrazin-2(1H)-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1OSR
 
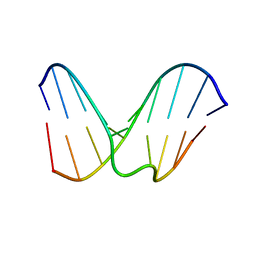 | | Structural study of dna duplex containaing a n-(2-deoxy-beta-erytho-pentofuranosyl) formamide frameshift by nmr and restrained molecular dynamics | | Descriptor: | 5'-D(*AP*GP*GP*AP*CP*CP*AP*CP*G)-3', 5'-D(*CP*GP*TP*GP*GP*(2DF)P*TP*CP*CP*T)-3' | | Authors: | Maufrais, C, Fazakerley, G.V, Cadet, J, Boulard, Y. | | Deposit date: | 2003-03-20 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural study of DNA duplex containing an N-(2-deoxy-beta-D-erythro-pentofuranosyl) formamide
frameshift by NMR and restrained molecular dynamics.
Nucleic Acids Res., 31, 2003
|
|
3OOL
 
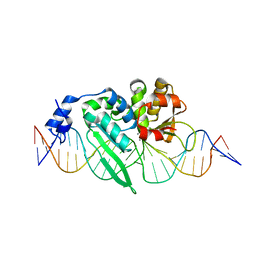 | | I-SceI complexed with C/G+4 DNA substrate | | Descriptor: | 5'-D(*CP*AP*CP*GP*CP*TP*AP*GP*GP*GP*AP*TP*AP*AP*CP*CP*GP*GP*GP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*GP*TP*AP*TP*TP*AP*CP*CP*CP*GP*GP*TP*TP*AP*TP*CP*CP*CP*TP*AP*GP*CP*GP*T)-3', CALCIUM ION, ... | | Authors: | Joshi, R, Chen, J.-H, Golden, B.L, Gimble, F.S. | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of I-SceI Homing Endonucleases with Increased DNA Recognition Site Specificity.
J.Mol.Biol., 405, 2011
|
|
