2L33
 
 | | Solution NMR Structure of DRBM 2 domain of Interleukin enhancer-binding factor 3 from Homo sapiens, Northeast Structural Genomics Consortium Target HR4527E | | Descriptor: | Interleukin enhancer-binding factor 3 | | Authors: | Liu, G, Janjua, H, Xiao, R, Acton, T.B, Ciccosanti, A, Shastry, R.B, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-09-03 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR4527E
To be Published
|
|
2B8A
 
 | | High Resolution Structure of the HDGF PWWP Domain | | Descriptor: | Hepatoma-derived growth factor | | Authors: | Lukasik, S.M, Cierpicki, T, Borloz, M, Grembecka, J, Everett, A, Bushweller, J.H. | | Deposit date: | 2005-10-06 | | Release date: | 2005-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of the HDGF PWWP domain: a potential DNA binding domain.
Protein Sci., 15, 2006
|
|
2LPE
 
 | |
2LT1
 
 | |
2LQA
 
 | |
2MO7
 
 | |
2MO2
 
 | |
2LZI
 
 | |
2MFS
 
 | |
2M1B
 
 | |
2LZY
 
 | |
2LXF
 
 | |
2MCO
 
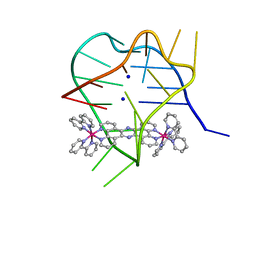 | | Structural studies on dinuclear ruthenium(II) complexes that bind diastereoselectively to an anti-parallel folded human telomere sequence | | Descriptor: | SODIUM ION, human telomere quadruplex, tetrakis(2,2'-bipyridine-kappa~2~N~1~,N~1'~)(mu-tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~13~,N~14~)diruthenium(4+) L enantiomer | | Authors: | Williamson, M.P, Wilson, T, Thomas, J.A, Felix, V, Costa, P.J. | | Deposit date: | 2013-08-22 | | Release date: | 2013-10-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Studies on Dinuclear Ruthenium(II) Complexes That Bind Diastereoselectively to an Antiparallel Folded Human Telomere Sequence.
J.Med.Chem., 56, 2013
|
|
2M1A
 
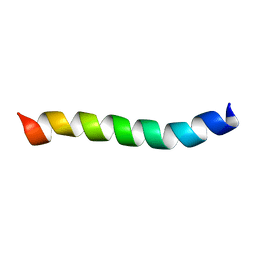 | | HIV-1 Rev ARM peptide (residues T34-R50) | | Descriptor: | HIV-1 Rev arginine-rich motif (ARM) | | Authors: | Casu, F, Duggan, B.M, Hennig, M. | | Deposit date: | 2012-11-21 | | Release date: | 2013-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Arginine-Rich RNA-Binding Motif of HIV-1 Rev Is Intrinsically Disordered and Folds upon RRE Binding.
Biophys.J., 105, 2013
|
|
2LZX
 
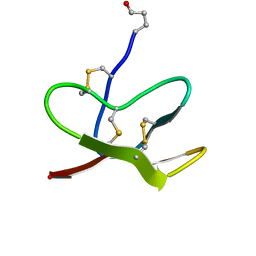 | |
2M8N
 
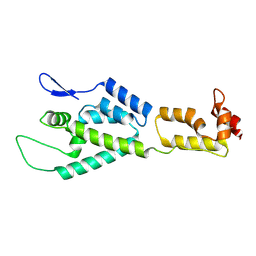 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2N3P
 
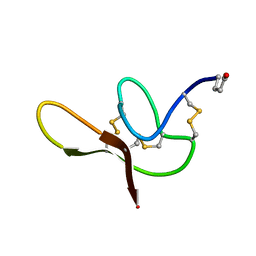 | |
2MS0
 
 | |
1XFR
 
 | |
2N24
 
 | |
2N2G
 
 | |
2MS1
 
 | |
2MQT
 
 | |
1YUI
 
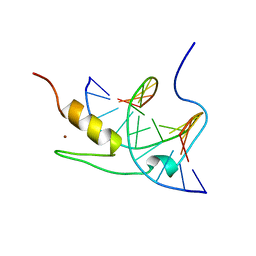 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
1YUJ
 
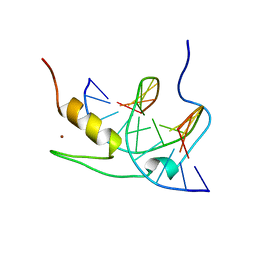 | | SOLUTION NMR STRUCTURE OF THE GAGA FACTOR/DNA COMPLEX, 50 STRUCTURES | | Descriptor: | DNA (5'-D(*GP*CP*CP*GP*AP*GP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*CP*TP*CP*TP*CP*GP*GP*C)-3'), GAGA-FACTOR, ... | | Authors: | Clore, G.M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1996-12-31 | | Release date: | 1997-12-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a specific GAGA factor-DNA complex reveals a modular binding mode.
Nat.Struct.Biol., 4, 1997
|
|
