5ZRU
 
 | | Crystal structure of Agl-KA catalytic domain | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alpha-1,3-glucanase, CALCIUM ION, ... | | Authors: | Yano, S, Makabe, K. | | Deposit date: | 2018-04-25 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | Crystal structure of the catalytic unit of GH 87-type alpha-1,3-glucanase Agl-KA from Bacillus circulans.
Sci Rep, 9, 2019
|
|
5GVA
 
 | | WD40 domain of human AND-1 | | Descriptor: | WD repeat and HMG-box DNA-binding protein 1 | | Authors: | Guan, C.C, Li, J. | | Deposit date: | 2016-09-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | The structure and polymerase-recognition mechanism of the crucial adaptor protein AND-1 in the human replisome
J. Biol. Chem., 292, 2017
|
|
5TEX
 
 | | Pim-1 kinase in complex with a 7-azaindole | | Descriptor: | (4-{4-chloro-1-methyl-2-[4-(piperazin-1-yl)phenyl]-1H-pyrrolo[2,3-b]pyridin-3-yl}phenyl)methanol, IMIDAZOLE, Serine/threonine-protein kinase pim-1 | | Authors: | Mechin, I, Wang, R, Batchelor, J.D, McLean, L. | | Deposit date: | 2016-09-23 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7TC0
 
 | | The structure of human ABCA1 in digitonin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
5GH0
 
 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
7TBY
 
 | | The structure of human ABCA1 in nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
7TBW
 
 | | The structure of ATP-bound ABCA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
7TBZ
 
 | | The structure of ABCA1 Y482C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATP-binding cassette, ... | | Authors: | Sun, Y, Li, X. | | Deposit date: | 2021-12-22 | | Release date: | 2022-01-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cholesterol efflux mechanism revealed by structural analysis of human ABCA1 conformational states.
Nat Cardiovasc Res, 1, 2022
|
|
5T7G
 
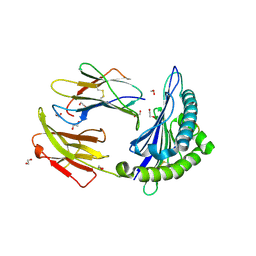 | | Crystal Structure of Murine MHC-I H-2Dd in complex with Murine Beta2-Microglobulin and a Variant of Peptide (PT9) of HIV gp120 MN Isolate (IGPGRAFYT) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J, Natarajan, K, Margulies, D. | | Deposit date: | 2016-09-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Effects of Cross-Presentation, Antigen Processing, and Peptide Binding in HIV Evasion of T Cell Immunity.
J. Immunol., 200, 2018
|
|
2R46
 
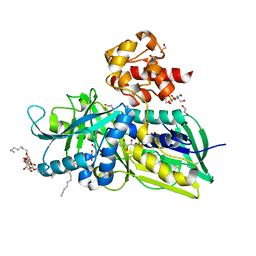 | | Crystal structure of Escherichia coli Glycerol-3-phosphate Dehydrogenase in complex with 2-phosphopyruvic acid. | | Descriptor: | 1,2-ETHANEDIOL, Aerobic glycerol-3-phosphate dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yeh, J.I, Du, S, Chinte, U. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of glycerol-3-phosphate dehydrogenase, an essential
monotopic membrane enzyme involved in respiration and metabolism
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
7X2M
 
 | | Crystal structure of nanobody 1-2C7 with SARS-CoV-2 RBD | | Descriptor: | 1-2C7, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | Authors: | Wang, X.Q, Zhang, L.Q, Ren, Y.F, Li, M.X. | | Deposit date: | 2022-02-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Broadly neutralizing and protective nanobodies against SARS-CoV-2 Omicron subvariants BA.1, BA.2, and BA.4/5 and diverse sarbecoviruses.
Nat Commun, 13, 2022
|
|
4MOW
 
 | |
9FBF
 
 | | VDR complex with UG-481 | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(1~{S})-5-methyl-1-[(1~{R},2~{R})-2-(3-methyl-3-oxidanyl-butyl)cyclopropyl]-5-oxidanyl-hexyl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2024-05-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of New Type of Gemini Analogues with a Cyclopropane Moiety in Their Side Chain.
J.Med.Chem., 67, 2024
|
|
1WZ3
 
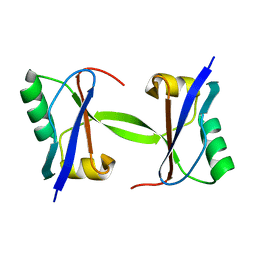 | | The crystal structure of plant ATG12 | | Descriptor: | autophagy 12b | | Authors: | Suzuki, N.N, Yoshimoto, K, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-06-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of plant ATG12 and its biological implication in autophagy.
Autophagy, 1, 2005
|
|
3X2G
 
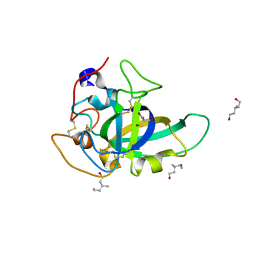 | | X-ray structure of PcCel45A N92D apo form at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
3KW4
 
 | | Crystal structure of cytochrome 2B4 in complex with the anti-platelet drug ticlopidine | | Descriptor: | 2-{[(3alpha,5alpha,7alpha,8alpha,10alpha,12alpha,17alpha)-3,12-bis{2-[(4-O-alpha-D-glucopyranosyl-beta-D-glucopyranosyl)oxy]ethoxy}cholan-7-yl]oxy}ethyl 4-O-alpha-D-glucopyranosyl-beta-D-glucopyranoside, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Gay, S.C, Maekawa, K, Roberts, A.G, Hong, W.-X, Zhang, Q, Stout, C.D, Halpert, J.R. | | Deposit date: | 2009-11-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structures of cytochrome P450 2B4 complexed with the antiplatelet drugs ticlopidine and clopidogrel.
Biochemistry, 49, 2010
|
|
5TRO
 
 | | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 1 | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dimerization and Transpeptidase domains (residues 39-608) of Penicillin-Binding Protein 1 from Staphylococcus aureus.
To Be Published
|
|
5TSV
 
 | |
3X2L
 
 | | X-ray structure of PcCel45A apo form at 95K. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-methylpentane-1,5-diol, Endoglucanase V-like protein | | Authors: | Nakamura, A, Ishida, T, Ohta, K, Tanaka, H, Inaka, K, Samejima, M, Igarashi, K. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
5KOK
 
 | | Pavine N-methyltransferase in complex with Tetrahydropapaverine and S-adenosylhomocysteine pH 7.25 | | Descriptor: | (1~{R})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pavine N-methyltransferase, ... | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
6ATO
 
 | |
5J1E
 
 | | Crystal Structure of a Hydroxypyridone Carboxylic Acid Active-Site RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 5-hydroxy-4-oxo-1-[(4'-sulfamoyl[1,1'-biphenyl]-4-yl)methyl]-1,4-dihydropyridine-3-carboxylic acid, HIV-1 REVERSE TRANSCRIPTASE P51 DOMAIN, HIV-1 REVERSE TRANSCRIPTASE P66 DOMAIN, ... | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-03-29 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H.
J.Med.Chem., 59, 2016
|
|
4YDD
 
 | | Crystal structure of the perchlorate reductase PcrAB from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tsai, C.-L, Youngblut, M.D, Tainer, J.A. | | Deposit date: | 2015-02-21 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
5T6L
 
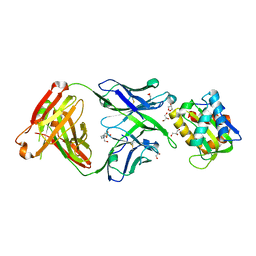 | | Crystal structure of 10E8 Fab in complex with the MPER epitope scaffold T117v2 | | Descriptor: | 10E8 EPITOPE SCAFFOLD T117V2, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
