3E42
 
 | | Q138F HincII bound to GTCGAC and Ca2+ (cocrystallized) | | Descriptor: | 5'-D(*DGP*DCP*DCP*DGP*DGP*DTP*DCP*DGP*DAP*DCP*DCP*DGP*DGP*DC)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Horton, N.C, Babic, A.C, Little, E.J, Manohar, V.M. | | Deposit date: | 2008-08-08 | | Release date: | 2008-08-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | DNA distortion and specificity in a sequence-specific endonuclease.
J.Mol.Biol., 383, 2008
|
|
4P3K
 
 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
3FYH
 
 | | Recombinase in complex with ADP and metatungstate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA repair and recombination protein radA, MAGNESIUM ION, ... | | Authors: | Li, Y, He, Y, Luo, Y. | | Deposit date: | 2009-01-22 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal Rad51 homologue in complex with a metatungstate inhibitor.
Biochemistry, 48, 2009
|
|
2W1S
 
 | |
3G1N
 
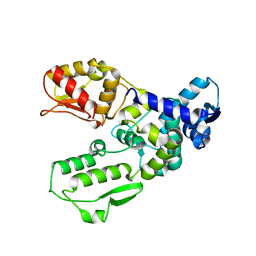 | | Catalytic domain of the human E3 ubiquitin-protein ligase HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, SODIUM ION | | Authors: | Walker, J.R, Qiu, L, Li, Y, Davis, T, Tempel, W, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Botchkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-01-30 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hect Domain of Human HUWE1/MULE
To be Published
|
|
4OQ4
 
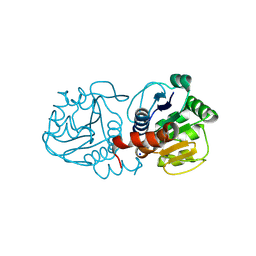 | | Crystal Structure of E18A Human DJ-1 | | Descriptor: | Protein DJ-1, SODIUM ION | | Authors: | Prahlad, J, Hauser, D.N, Milkovic, N.M, Cookson, M.R, Wilson, M.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Use of cysteine-reactive cross-linkers to probe conformational flexibility of human DJ-1 demonstrates that Glu18 mutations are dimers.
J Neurochem, 130, 2014
|
|
3FZQ
 
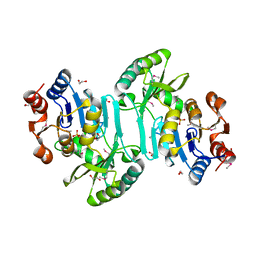 | |
4OWE
 
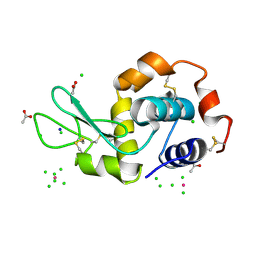 | | PtCl6 binding to HEWL | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Starkey, V.L, Lamplough, L, Kaenket, S, Helliwell, J.R. | | Deposit date: | 2014-01-31 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The binding of platinum hexahalides (Cl, Br and I) to hen egg-white lysozyme and the chemical transformation of the PtI6 octahedral complex to a PtI3 moiety bound to His15.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3CYM
 
 | | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis | | Descriptor: | GLYCEROL, SODIUM ION, Uncharacterized protein BAD_0989 | | Authors: | Patskovsky, Y, Ozyurt, S, Freeman, J, Chang, S, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of protein BAD_0989 from Bifidobacterium adolescentis.
To be Published
|
|
4PIG
 
 | | Crystal structure of the ubiquitin K11S mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Loll, P.J, Xu, P.J, Schmidt, J, Melideo, S.L. | | Deposit date: | 2014-05-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Enhancing ubiquitin crystallization through surface-entropy reduction.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4PO9
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-BR | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4POD
 
 | | Structure of Triosephosphate Isomerase I170V mutant human enzyme. | | Descriptor: | BROMIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Amrich, C.G, Aslam, A.A, Heroux, A, VanDemark, A.P. | | Deposit date: | 2014-02-25 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Triosephosphate isomerase I170V alters catalytic site, enhances stability and induces pathology in a Drosophila model of TPI deficiency.
Biochim.Biophys.Acta, 1852, 2015
|
|
2X7R
 
 | |
4PUV
 
 | | URATE OXIDASE DI-AZIDE complex | | Descriptor: | AZIDE ION, SODIUM ION, Uricase | | Authors: | Colloc'h, N, Prange, T. | | Deposit date: | 2014-03-14 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Azide inhibition of urate oxidase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1B7R
 
 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1999-01-25 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
4PIU
 
 | |
4PJ8
 
 | | Structure of human MR1-5-OP-RU in complex with human MAIT TRBV20 TCR | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Birkinshaw, R.W, Rossjohn, J. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular basis underpinning the T cell receptor heterogeneity of mucosal-associated invariant T cells.
J.Exp.Med., 211, 2014
|
|
3GED
 
 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | Descriptor: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
4TTT
 
 | |
4TR8
 
 | | Crystal structure of DNA polymerase sliding clamp from Pseudomonas aeruginosa | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Olieric, V, Burnouf, D, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
3FVU
 
 | | Crystal Structure of Human Kynurenine Aminotransferase I in Complex with Indole-3-acetic Acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, GLYCEROL, Kynurenine--oxoglutarate transaminase 1, ... | | Authors: | Han, Q, Robinson, H, Cai, T, Tagle, D.A, Li, J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into the inhibition of human kynurenine aminotransferase I/glutamine transaminase K
J.Med.Chem., 52, 2009
|
|
2WD0
 
 | | CRYSTAL STRUCTURE OF NONSYNDROMIC DEAFNESS (DFNB12) ASSOCIATED MUTANT D124G OF MOUSE CADHERIN-23 EC1-2 | | Descriptor: | CADHERIN-23, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2009-03-18 | | Release date: | 2010-04-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Determinants of Cadherin-23 Function in Hearing and Deafness.
Neuron, 66, 2010
|
|
3G3K
 
 | |
1B5X
 
 | | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and x-ray analysis of six ser->ala mutants | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
2VWM
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | (4R)-4-{[(5-chlorothiophen-2-yl)carbonyl]amino}-N-(cyclopropylmethyl)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-L-prolinamide, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
