2PIZ
 
 | |
2R0G
 
 | | Chromopyrrolic acid-soaked RebC with bound 7-carboxy-K252c | | Descriptor: | 7-carboxy-5-hydroxy-12,13-dihydro-6H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, FLAVIN-ADENINE DINUCLEOTIDE, RebC | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2O48
 
 | | Crystal structure of Mammalian Dimeric Dihydrodiol Dehydrogenase | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, P-HYDROXYACETOPHENONE, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2006-12-04 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of dimeric dihydrodiol dehydrogenase apoenzyme and inhibitor complex: probing the subunit interface with site-directed mutagenesis.
Proteins, 70, 2008
|
|
3CYZ
 
 | | Dimeric crystal structure of a pheromone binding protein from Apis mellifera in complex with 9-keto-2(E)-decenoic acid at pH 7.0 | | Descriptor: | (2Z)-9-oxodec-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pesenti, M.E, Spinelli, S, Bezirard, V, Briand, L, Pernollet, J.C, Tegoni, M, Cambillau, C. | | Deposit date: | 2008-04-27 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Queen bee pheromone binding protein pH-induced domain swapping favors pheromone release
J.Mol.Biol., 390, 2009
|
|
2OWO
 
 | | Last Stop on the Road to Repair: Structure of E.coli DNA Ligase Bound to Nicked DNA-Adenylate | | Descriptor: | 26-MER, 5'-D(*AP*CP*AP*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*AP*TP*G)-3', ... | | Authors: | Shuman, S, Nandakumar, J, Nair, P.A. | | Deposit date: | 2007-02-16 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Last Stop on the Road to Repair: Structure of E. coli DNA Ligase Bound to Nicked DNA-Adenylate.
Mol.Cell, 26, 2007
|
|
2R0P
 
 | | K252c-soaked RebC | | Descriptor: | 6,7,12,13-tetrahydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazol-5-one, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic trapping in the rebeccamycin biosynthetic enzyme RebC
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R0C
 
 | |
2R6K
 
 | | Crystal structure of an I71V hGSTA1-1 mutant in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Dirr, H.W, Fisher, L, Burke, J.P.W.G, Sayed, M, Sewell, T. | | Deposit date: | 2007-09-06 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4BU7
 
 | | Crystal structure of human tankyrase 2 in complex with 2-(4- bromophenyl)-3,4-dihydroquinazolin-4-one | | Descriptor: | 2-(4-bromophenyl)-3,4-dihydroquinazolin-4-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
4XA7
 
 | | Soluble part of holo NqrC from V. harveyi | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit C | | Authors: | Borshchevskiy, V, Round, E, Bertsova, Y, Polovinkin, V, Gushchin, I, Mishin, A, Kovalev, K, Kachalova, G, Popov, A, Bogachev, A, Gordeliy, V. | | Deposit date: | 2014-12-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Investigation of Flavin Binding Center of the NqrC Subunit of Sodium-Translocating NADH:Quinone Oxidoreductase from Vibrio harveyi.
Plos One, 10, 2015
|
|
4X9V
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3C | | Descriptor: | Phosphopeptide macrocycle 3C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4BUE
 
 | | Crystal structure of human tankyrase 2 in complex with 3-methyl-N-(4-(4-oxo-3,4-dihydroquinazolin-2-yl)phenyl)butanamide | | Descriptor: | 3-METHYL-N-[4-(4-OXO-3,4-DIHYDROQUINAZOLIN-2-YL)PHENYL)BUTANAMIDE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Para-Substituted 2-Phenyl-3,4-Dihydroquinazolin-4-Ones as Potent and Selective Tankyrase Inhibitors.
Chemmedchem, 8, 2013
|
|
4XEJ
 
 | | IRES bound to bacterial Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Zhu, J, Korostelev, A, Noller, H.F, Donohue, J.P. | | Deposit date: | 2014-12-23 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Initiation of translation in bacteria by a structured eukaryotic IRES RNA.
Nature, 519, 2015
|
|
4CIL
 
 | |
3LAD
 
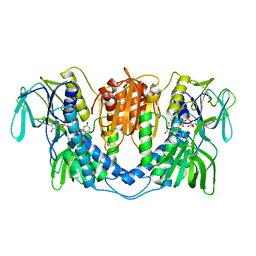 | |
4CXR
 
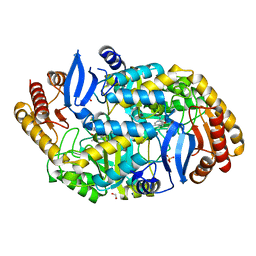 | | Mycobaterium tuberculosis transaminase BioA complexed with 1-(1,3- benzothiazol-2-yl)methanamine | | Descriptor: | 1,2-ETHANEDIOL, 1-(1,3-benzothiazol-2-yl)methanamine, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Geders, T.W, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
3LZO
 
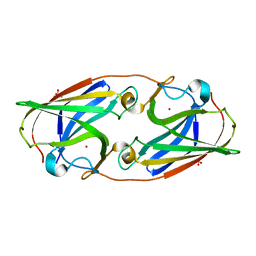 | | Crystal Structure Analysis of the copper-reconstituted P19 protein from Campylobacter jejuni at 1.65 A at pH 10.0 | | Descriptor: | COPPER (II) ION, P19 protein, SULFATE ION | | Authors: | Doukov, T.I, Chan, A.C.K, Scofield, M, Ramin, A.B, Tom-Yew, S.A.L, Murphy, M.E.P. | | Deposit date: | 2010-03-01 | | Release date: | 2010-07-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of P19, a High-Affinity Iron Transporter of the Human Pathogen Campylobacter jejuni.
J.Mol.Biol., 401, 2010
|
|
4X9W
 
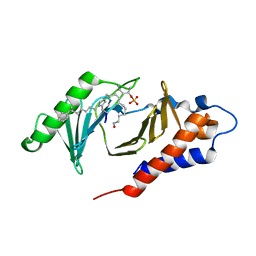 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 4C | | Descriptor: | Macrocyclic phosphopeptide 4C, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
4CXQ
 
 | | Mycobaterium tuberculosis transaminase BioA complexed with substrate KAPA | | Descriptor: | 1,2-ETHANEDIOL, 7-KETO-8-AMINOPELARGONIC ACID, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Wilson, D.J, Geders, T.W, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
3V8E
 
 | | Crystal structure of the yeast nicotinamidase Pnc1p bound to the inhibitor nicotinaldehyde | | Descriptor: | MAGNESIUM ION, Nicotinamidase, ZINC ION | | Authors: | Hoadley, K.A, Smith, B.C, Denu, J.M, Keck, J.L. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-25 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural and Kinetic Isotope Effect Studies of Nicotinamidase (Pnc1) from Saccharomyces cerevisiae.
Biochemistry, 51, 2012
|
|
4CQE
 
 | | B-Raf Kinase V600E mutant in complex with a diarylthiazole B-Raf Inhibitor | | Descriptor: | N-{4-[2-(1-cyclopropylpiperidin-4-yl)-4-(3-{[(2,5-difluorophenyl)sulfonyl]amino}-2-fluorophenyl)-1,3-thiazol-5-yl]pyridin-2-yl}acetamide, SLC45A3-BRAF FUSION PROTEIN | | Authors: | Casale, E, Fasolini, M, Pulici, M, Traquandi, G, Marchionni, C, Modugno, M, Lupi, R, Amboldi, N, Colombo, N, Corti, L, Gasparri, F, Pastori, W, Scolaro, A, Donati, D, Felder, E, Galvani, A, Isacchi, A, Pesenti, E, Ciomei, M. | | Deposit date: | 2014-02-14 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Optimization of Diarylthiazole B-Raf Inhibitors: Identification of a Compound Endowed with High Oral Antitumor Activity, Mitigated Herg Inhibition, and Low Paradoxical Effect.
Chemmedchem, 10, 2015
|
|
3ET0
 
 | |
6IJF
 
 | | Crystal structure of the type VI effector-immunity complex (Tae4-Tai4) from Agrobacterium tumefaciens | | Descriptor: | PENTAETHYLENE GLYCOL, SULFATE ION, Tae4, ... | | Authors: | Fukuhara, S, Nakane, T, Yamashita, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2018-10-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Agrobacterium tumefaciens type VI effector-immunity complex.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4X9R
 
 | | PLK-1 polo-box domain in complex with Bioactive Imidazolium-containing phosphopeptide macrocycle 3B | | Descriptor: | Phosphopeptide macrocycle 3B, Serine/threonine-protein kinase PLK1 | | Authors: | Grant, R.A, Qian, W.-J, Yaffe, M.B, Burke, T.R. | | Deposit date: | 2014-12-11 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Neighbor-directed histidine N ( tau )-alkylation: A route to imidazolium-containing phosphopeptide macrocycles.
Biopolymers, 104, 2015
|
|
6IU8
 
 | | Crystal structure of cytoplasmic metal binding domain with cobalt ions | | Descriptor: | COBALT (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
