3PF0
 
 | |
3PQA
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661 | | Descriptor: | Lactaldehyde dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase GapN from Methanocaldococcus jannaschii DSM 2661
To be Published
|
|
3PRB
 
 | |
3PF4
 
 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PVG
 
 | | Crystal structure of Z. mays CK2 alpha subunit in complex with the inhibitor 4,5,6,7-tetrabromo-1-carboxymethylbenzimidazole (K68) | | Descriptor: | (4,5,6,7-tetrabromo-1H-benzimidazol-1-yl)acetic acid, Casein kinase II subunit alpha | | Authors: | Papinutto, E, Franchin, C, Battistutta, R. | | Deposit date: | 2010-12-07 | | Release date: | 2010-12-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ATP site-directed inhibitors of protein kinase CK2: an update.
Curr Top Med Chem, 11, 2011
|
|
2HX5
 
 | |
3PFE
 
 | |
3PFU
 
 | | N-terminal domain of Thiol:disulfide interchange protein DsbD in its reduced form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thiol:disulfide interchange protein dsbD | | Authors: | Mavridou, D.A.I, Saridakis, E, Ferguson, S.J, Redfield, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxidation state-dependent protein-protein interactions in disulfide cascades
J.Biol.Chem., 286, 2011
|
|
3PFS
 
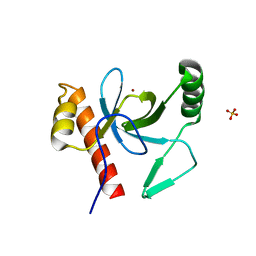 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 3 | | Descriptor: | Bromodomain and PHD finger-containing protein 3, SULFATE ION, ZINC ION | | Authors: | Lam, R, Zeng, H, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3Q3N
 
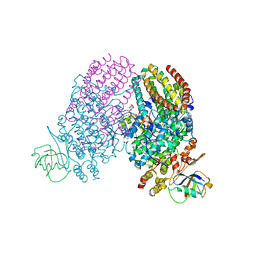 | | Toluene 4 monooxygenase HD complex with p-nitrophenol | | Descriptor: | FE (III) ION, P-NITROPHENOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Acheson, J.F, Bailey, L.J, Fox, B.G. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystallographic analysis of active site contributions to regiospecificity in the diiron enzyme toluene 4-monooxygenase.
Biochemistry, 51, 2012
|
|
3Q4F
 
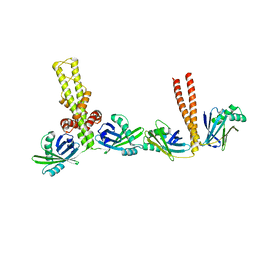 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
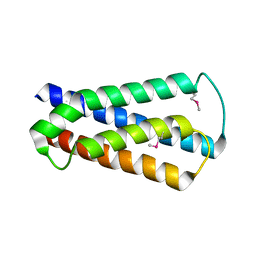
2HUJ
 
 | |
2HX1
 
 | |
3PFD
 
 | |
3PFO
 
 | |
2ANJ
 
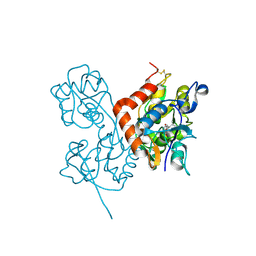 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
3PKV
 
 | |
3PM9
 
 | |
3PG3
 
 | | Human p38 MAP Kinase in Complex with RL182 | | Descriptor: | 1-[3-tert-butyl-1-(4-methylphenyl)-1H-pyrazol-5-yl]-3-{4-[2-(pyridin-3-ylmethoxy)ethyl]-1,3-thiazol-2-yl}urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Getlik, M, Rauh, D. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design and synthesis of cell active N-pyrazole, N-thiazole urea inhibitors of the MAP kinase p38alpha
To be Published
|
|
3PNX
 
 | |
3POQ
 
 | |
2AQJ
 
 | | The structure of tryptophan 7-halogenase (PrnA) suggests a mechanism for regioselective chlorination | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, TRYPTOPHAN, ... | | Authors: | Dong, C, Flecks, S, Unversucht, S, Haupt, C, Van Pee, K.-H, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2005-08-18 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Science, 309, 2005
|
|
2ATB
 
 | | Triple mutant 8D9D10V of scorpion toxin LQH-alpha-IT | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NITRATE ION, ... | | Authors: | Kahn, R, Karbat, I, Gurevitz, M, Frolow, F. | | Deposit date: | 2005-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
To be Published
|
|
3PQU
 
 | |
3PVY
 
 | |
