1UH8
 
 | |
5BK6
 
 | | Structural and biochemical characterization of a non-canonical biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Putative amidase | | Authors: | Peat, T.S, Esquirol, L, Newman, J, Scott, C. | | Deposit date: | 2017-09-12 | | Release date: | 2018-02-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural and biochemical characterization of the biuret hydrolase (BiuH) from the cyanuric acid catabolism pathway of Rhizobium leguminasorum bv. viciae 3841.
PLoS ONE, 13, 2018
|
|
2VQ4
 
 | |
6SK6
 
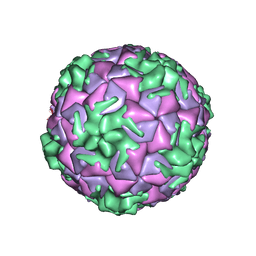 | | Cryo-EM structure of rhinovirus-B5 | | Descriptor: | Rhinovirus B5 VP1, Rhinovirus B5 VP2, Rhinovirus B5 VP3, ... | | Authors: | Wald, J, Goessweiner-Mohr, N, Blaas, D, Pasin, M. | | Deposit date: | 2019-08-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of pleconaril-resistant rhinovirus-B5 complexed to the antiviral OBR-5-340 reveals unexpected binding site.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2V8M
 
 | |
1AYM
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
1AYN
 
 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
9D8A
 
 | |
2VQR
 
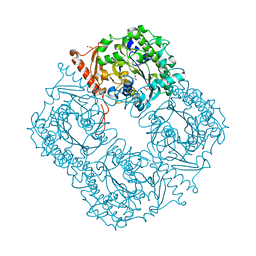 | | Crystal structure of a phosphonate monoester hydrolase from rhizobium leguminosarum: a new member of the alkaline phosphatase superfamily | | Descriptor: | ACETATE ION, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Jonas, S, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2008-03-18 | | Release date: | 2008-09-30 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A New Member of the Alkaline Phosphatase Superfamily with a Formylglycine Nucleophile: Structural and Kinetic Characterisation of a Phosphonate Monoester Hydrolase/Phosphodiesterase from Rhizobium Leguminosarum.
J.Mol.Biol., 384, 2008
|
|
1X3U
 
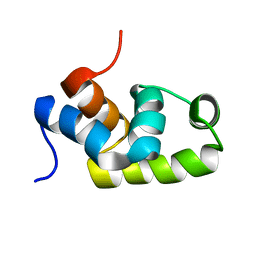 | | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium melilot | | Descriptor: | Transcriptional regulatory protein fixJ | | Authors: | Kurashima-Ito, K, Kasai, Y, Hosono, K, Tamura, K, Oue, S, Isogai, M, Ito, Y, Nakamura, H, Shiro, Y. | | Deposit date: | 2005-05-10 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal transcriptional activator domain of FixJ from Sinorhizobium meliloti and its recognition of the fixK promoter
Biochemistry, 44, 2005
|
|
5FX6
 
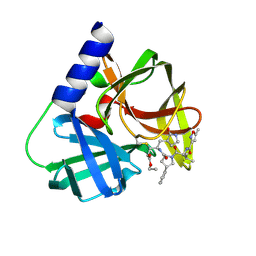 | | Novel inhibitors of human rhinovirus 3C protease | | Descriptor: | RHINOVIRUS 3C PROTEASE, ethyl (4R)-4-[[(2S,4S)-1-[(2S)-3-methyl-2-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]butanoyl]-4-phenyl-pyrrolidin-2-yl]carbonylamino]-5-[(3S)-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Kawatkar, S, Ek, M, Hoesch, V, Gagnon, M, Nilsson, E, Lister, T, Olsson, L, Patel, J, Yu, Q. | | Deposit date: | 2016-02-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design and Structure-Activity Relationships of Novel Inhibitors of Human Rhinovirus 3C Protease.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
6IZ9
 
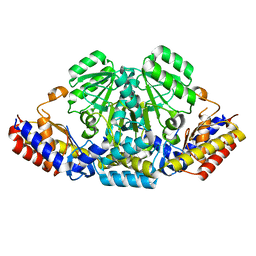 | |
6DE6
 
 | |
1BOL
 
 | |
6QPP
 
 | | Rhizomucor miehei lipase propeptide complex, native | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
4OMY
 
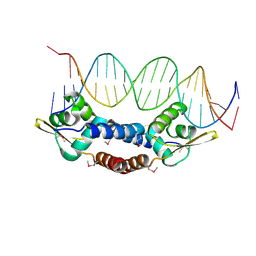 | |
4OMZ
 
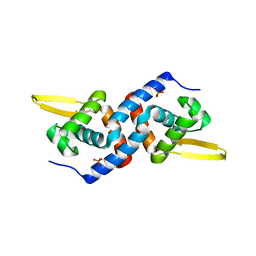 | |
4ON0
 
 | | Crystal Structure of NolR from Sinorhizobium fredii in complex with oligo AA DNA | | Descriptor: | DNA (5 -D(*TP*AP*AP*TP*CP*TP*CP*TP*TP*GP*GP*GP*AP*CP*TP*AP*CP*AP*AP*TP*TP*A)-3 ), DNA (5 -D(*TP*AP*TP*TP*AP*GP*AP*GP*AP*AP*CP*CP*CP*TP*GP*AP*TP*GP*TP*TP*AP*A)-3 ), NolR | | Authors: | Lee, S.G, Krishnan, H.B, Jez, J.M. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for regulation of rhizobial nodulation and symbiosis gene expression by the regulatory protein NolR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NV8
 
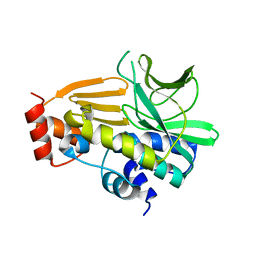 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase F42W Mutant | | Descriptor: | Arylamine N-acetyltransferase | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4NV7
 
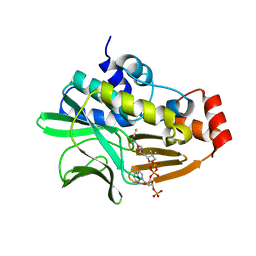 | | Crystal Structure of Mesorhizobium Loti Arylamine N-acetyltransferase 1 In Complex With CoA | | Descriptor: | Arylamine N-acetyltransferase, COENZYME A | | Authors: | Xu, X.M, Haouz, A, Weber, P, Li de la sierra-gallay, I, Kubiak, X, Dupret, J.-M, Rodrigues-lima, F. | | Deposit date: | 2013-12-05 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insight into cofactor recognition in arylamine N-acetyltransferase enzymes: structure of Mesorhizobium loti arylamine N-acetyltransferase in complex with coenzyme A.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1RUC
 
 | | RHINOVIRUS 14 MUTANT N1105S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUI
 
 | | RHINOVIRUS 14 MUTANT S1223G COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52084 | | Descriptor: | 5-(7-(5-HYDRO-4-METHYL-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUE
 
 | | RHINOVIRUS 14 SITE DIRECTED MUTANT N1219A COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
1RUG
 
 | | RHINOVIRUS 14 MUTANT N1219S COMPLEXED WITH ANTIVIRAL COMPOUND WIN 52035 | | Descriptor: | 5-(5-(4-(4,5-DIHYDRO-2-OXAZOLY)PHENOXY)PENTYL)-3-METHYL ISOXAZOLE, RHINOVIRUS 14 | | Authors: | Hadfield, A, Oliveira, M.A, Kim, K.H, Minor, I, Kremer, M.J, Heinz, B.A, Shepard, D, Pevear, D.C, Rueckert, R.R, Rossmann, M.G. | | Deposit date: | 1995-06-09 | | Release date: | 1995-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on human rhinovirus 14 drug-resistant compensation mutants.
J.Mol.Biol., 253, 1995
|
|
