2VQA
 
 | | Protein-folding location can regulate Mn versus Cu- or Zn-binding. Crystal Structure of MncA. | | Descriptor: | ACETATE ION, MANGANESE (II) ION, SLL1358 PROTEIN | | Authors: | Tottey, S, Waldron, K.J, Firbank, S.J, Reale, B, Bessant, C, Sato, K, Gray, J, Banfield, M.J, Dennison, C, Robinson, N.J. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Protein-Folding Location Can Regulate Manganese-Binding Versus Copper- or Zinc-Binding.
Nature, 455, 2008
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UY9
 
 | | E162A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
1CAW
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-06-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CAX
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-06-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CAV
 
 | |
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
1CAU
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-07-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2UYA
 
 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
1DGR
 
 | |
1DGW
 
 | |
2D5F
 
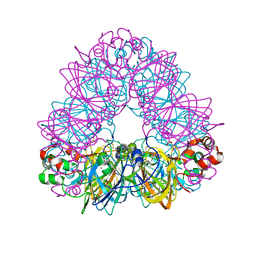 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2EA7
 
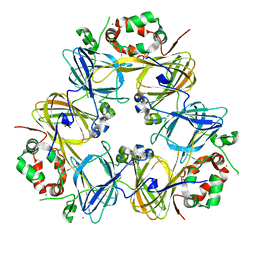 | | Crystal Structure of Adzuki Bean 7S Globulin-1 | | Descriptor: | 7S globulin-1, ACETIC ACID, CALCIUM ION | | Authors: | Fukuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystallography of recombinant 7S globulins of Adzuki bean and structure-function relationships with 7S globulins of various crops.
J.Agric.Food Chem., 56, 2008
|
|
2EAA
 
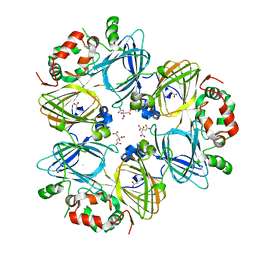 | | Crystal Structure of Adzuki Bean 7S Globulin-3 | | Descriptor: | 7S globulin-3, ACETIC ACID, CALCIUM ION, ... | | Authors: | Fukuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and crystallography of recombinant 7S globulins of Adzuki bean and structure-function relationships with 7S globulins of various crops.
J.Agric.Food Chem., 56, 2008
|
|
2E9Q
 
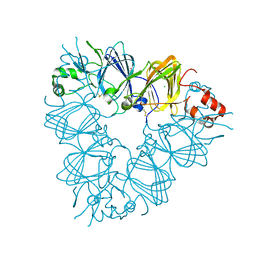 | | Recombinant pro-11S globulin of pumpkin | | Descriptor: | 11S globulin subunit beta, CHLORIDE ION, PHOSPHATE ION | | Authors: | Fukuda, T, Prak, K, Itoh, T, Masuda, T, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2007-01-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
2CAU
 
 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2CAV
 
 | | CANAVALIN FROM JACK BEAN | | Descriptor: | PROTEIN (CANAVALIN) | | Authors: | Ko, T.-P, Day, J, Macpherson, A. | | Deposit date: | 1998-11-20 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined structure of canavalin from jack bean in two crystal forms at 2.1 and 2.0 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2CV6
 
 | | Crystal Structure of 8Salpha Globulin, the Major Seed Storage Protein of Mungbean | | Descriptor: | Seed storage protein | | Authors: | Itoh, T, Garcia, R.N, Adachi, M, Maruyama, Y, Tecson-Mendoza, E.M, Mikami, B, Utsumi, S. | | Deposit date: | 2005-05-31 | | Release date: | 2006-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of 8Salpha globulin, the major seed storage protein of mung bean.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2ET1
 
 | | Oxalate oxidase in complex with substrate analogue glycolate | | Descriptor: | GLYOXYLIC ACID, MANGANESE (II) ION, Oxalate oxidase 1 | | Authors: | Rose, R.-S, Opaleye, O, Woo, E.-J, Pickersgill, R.W. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase
J.Biol.Chem., 281, 2006
|
|
2EVX
 
 | |
2ETE
 
 | | Recombinant oxalate oxidase in complex with glycolate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYOXYLIC ACID, MANGANESE (II) ION, ... | | Authors: | Opaleye, O, Rose, R.-S, Whittaker, M.M, Woo, E.-J, Whittaker, J.W, Pickersgill, R.W. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase
J.Biol.Chem., 281, 2006
|
|
2ET7
 
 | | Structural and spectroscopic insights into the mechanism of oxalate oxidase | | Descriptor: | MANGANESE (II) ION, Oxalate oxidase 1 | | Authors: | Opaleye, O, Rose, R.-S, Whittaker, M.M, Woo, E.-J, Whittaker, J.W, Pickersgill, R.W. | | Deposit date: | 2005-10-27 | | Release date: | 2005-11-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and spectroscopic studies shed light on the mechanism of oxalate oxidase
J.Biol.Chem., 281, 2006
|
|
2D5H
 
 | | Crystal Structure of Recombinant Soybean Proglycinin A3B4 subunit, its Comparison with Mature Glycinin A3B4 subunit, Responsible for Hexamer Assembly | | Descriptor: | CARBONATE ION, MAGNESIUM ION, glycinin A3B4 subunit | | Authors: | Itoh, T, Adachi, M, Masuda, T, Mikami, B, Utsumi, S. | | Deposit date: | 2005-11-01 | | Release date: | 2006-11-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 2010
|
|
