2FSO
 
 | |
5CV6
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/I92E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Theodoru, A, Robinson, A.C, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2015-07-25 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66H/I92E at cryogenic temperature
To be Published
|
|
1MEA
 
 | |
5VFR
 
 | | Nucleotide-driven Triple-state Remodeling of the AAA-ATPase Channel in the Activated Human 26S Proteasome | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 12, ... | | Authors: | Zhu, Y, Wang, W.L, Yu, D, Ouyang, Q, Lu, Y, Mao, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural mechanism for nucleotide-driven remodeling of the AAA-ATPase unfoldase in the activated human 26S proteasome.
Nat Commun, 9, 2018
|
|
7QHA
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol | | Descriptor: | Megabody c7HopQ, Putative TRAP-type C4-dicarboxylate transport system, large permease component, ... | | Authors: | North, R.A, Davies, J.S, Morado, D, Dobson, R.C.J. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
1MED
 
 | |
7QHJ
 
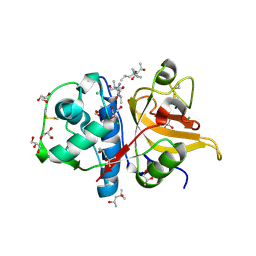 | | Peptide GAKSAA in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
1MPH
 
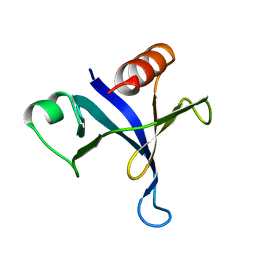 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
6KMM
 
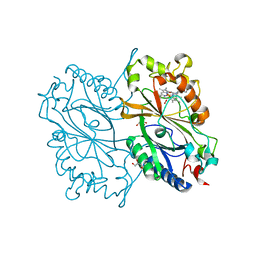 | | Crystal Structure of HEPES bound Dye Decolorizing peroxidase from Bacillus subtilis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dhankhar, P, Dalal, V, Mahto, J.K, Kumar, P. | | Deposit date: | 2019-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Characterization of dye-decolorizing peroxidase from Bacillus subtilis.
Arch.Biochem.Biophys., 693, 2020
|
|
1M2C
 
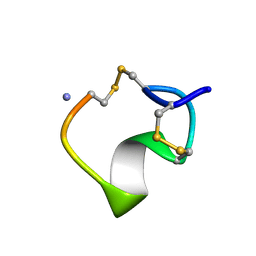 | | THREE-DIMENSIONAL STRUCTURE OF ALPHA-CONOTOXIN MII, NMR, 14 STRUCTURES | | Descriptor: | ALPHA-CONOTOXIN MII | | Authors: | Shon, K.J, Koerber, S.C, Rivier, J.E, Olivera, B.M, Mcintosh, J.M. | | Deposit date: | 1997-09-15 | | Release date: | 1998-12-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A new alpha-conotoxin which targets alpha3beta2 nicotinic acetylcholine receptors.
J.Biol.Chem., 271, 1996
|
|
7QDE
 
 | |
5CWC
 
 | |
5CWM
 
 | |
1MG7
 
 | | Crystal Structure of xol-1 | | Descriptor: | early switch protein xol-1 2.2k splice form | | Authors: | Luz, J.G, Hassig, C.A, Godzik, A, Meyer, B.J, Wilson, I.A. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators
Genes Dev., 17, 2003
|
|
7QFH
 
 | | Peptide AYFKKVL in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AMINO GROUP, CHLORIDE ION, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
1M0W
 
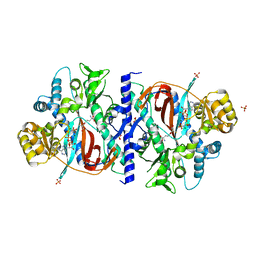 | | Yeast Glutathione Synthase Bound to gamma-glutamyl-cysteine, AMP-PNP and 2 Magnesium Ions | | Descriptor: | GAMMA-GLUTAMYLCYSTEINE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Gogos, A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-06-14 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large Conformational Changes in the Catalytic Cycle of Glutathione Synthase
Structure, 10, 2002
|
|
1MI3
 
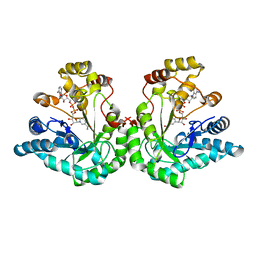 | | 1.8 Angstrom structure of xylose reductase from Candida tenuis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-08-21 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases
Biochem.J., 373, 2003
|
|
7QHK
 
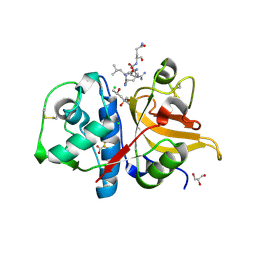 | | Peptide QLRQQE in complex with human cathepsin V C25A mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Proteomic data and structure analysis combined reveal interplay of structural rigidity and flexibility on selectivity of cysteine cathepsins.
Commun Biol, 6, 2023
|
|
2FOX
 
 | | CLOSTRIDIUM BEIJERINCKII FLAVODOXIN: SEMIQUINONE | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Ludwig, M.L, Pattridge, K.A, Metzger, A.L, Dixon, M.M, Eren, M, Feng, Y, Swenson, R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Control of oxidation-reduction potentials in flavodoxin from Clostridium beijerinckii: the role of conformation changes.
Biochemistry, 36, 1997
|
|
1MJ3
 
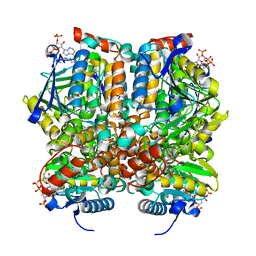 | | Crystal Structure Analysis of rat enoyl-CoA hydratase in complex with hexadienoyl-CoA | | Descriptor: | ENOYL-COA HYDRATASE, MITOCHONDRIAL, HEXANOYL-COENZYME A | | Authors: | Bell, A.F, Feng, Y, Hofstein, H.A, Parikh, S, Wu, J, Rudolph, M.J, Kisker, C, Tonge, P.J. | | Deposit date: | 2002-08-26 | | Release date: | 2002-09-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stereoselectivity of Enoyl-CoA Hydratase Results from Preferential Activation of
One of Two Bound Substrate Conformers
Chem.Biol., 9, 2002
|
|
7QUJ
 
 | | Structure of NsNEPS2, a 7S-cis-trans nepetalactone synthase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NsNEPS2 | | Authors: | Hernandez Lozada, N.J, Hong, B, Wood, J.C, Caputi, L, Basquin, J, Chuang, L, Kunert, M, Rodriguez Lopez, C.R, Langley, C, Zhao, D, Buell, C.R, Lichman, B.R, O'Connor, S.E. | | Deposit date: | 2022-01-18 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Biocatalytic routes to stereo-divergent iridoids.
Nat Commun, 13, 2022
|
|
1M24
 
 | |
4O5G
 
 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-5-[(4-aminophenyl)methylidene]-2-azanylidene-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
1MK5
 
 | | Wildtype Core-Streptavidin with Biotin at 1.4A. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Green, N.M, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
7QJR
 
 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
