1A3Q
 
 | | HUMAN NF-KAPPA-B P52 BOUND TO DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*GP*AP*TP*TP*CP*CP*CP*C)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B P52) | | Authors: | Cramer, P, Larson, C.J, Verdine, G.L, Muller, C.W. | | Deposit date: | 1998-01-23 | | Release date: | 1998-06-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution.
EMBO J., 16, 1997
|
|
6VTP
 
 | |
6VTS
 
 | |
6VTQ
 
 | |
6H5H
 
 | |
6VTR
 
 | | Crystal structure of G16S human Galectin-7 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Galectin-7 | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2020-02-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Perturbing dimer interactions and allosteric communication modulates the immunosuppressive activity of human galectin-7.
J.Biol.Chem., 297, 2021
|
|
6VTO
 
 | | Crystal structure of human Galectin-7 in complex with 4-O-beta-D-Galactopyranosyl-D-glucose | | Descriptor: | (2~{R},3~{R},4~{R},5~{R})-4-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2,3,5, 6-tetrakis(oxidanyl)hexanal, Galectin-7 | | Authors: | Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2020-02-13 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing dimer interactions and allosteric communication modulates the immunosuppressive activity of human galectin-7.
J.Biol.Chem., 297, 2021
|
|
5DVO
 
 | | Fc K392D/K409D homodimer | | Descriptor: | Ig gamma-1 chain C region, SULFATE ION | | Authors: | Atwell, S, Leaver-Fay, A, Froning, K.J, Aldaz, H, Pustilnik, A, Lu, F, Huang, F, Yuan, R, Dhanani, S.H, Chamberlain, A.K, Fitchett, J.R, Gutierrez, B, Hendle, J, Demarest, S.J, Kuhlman, B. | | Deposit date: | 2015-09-21 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computationally Designed Bispecific Antibodies using Negative State Repertoires.
Structure, 24, 2016
|
|
5GOX
 
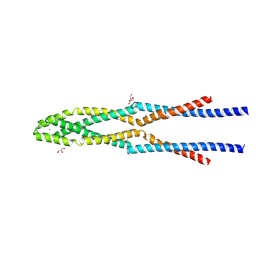 | | Eukaryotic Rad50 Functions as A Rod-shaped Dimer | | Descriptor: | DNA repair protein RAD50, GLYCEROL, ZINC ION | | Authors: | Park, Y.B, Hohl, M, Padjasek, M, Jeong, E, Jin, K.S, Krezel, A, Petrini, J.H.J, Cho, Y. | | Deposit date: | 2016-07-30 | | Release date: | 2017-02-01 | | Last modified: | 2017-03-15 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Eukaryotic Rad50 functions as a rod-shaped dimer
Nat. Struct. Mol. Biol., 24, 2017
|
|
4F92
 
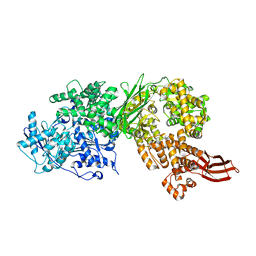 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8ZAK
 
 | |
8ZAH
 
 | |
9EUU
 
 | | Structure of recombinant alpha-synuclein fibrils 1B capable of seeding GCIs in vivo | | Descriptor: | Alpha-synuclein | | Authors: | Burger, D, Kashyrina, M, Lewis, A, De Nuccio, F, Mohammed, I, de La Seigliere, H, van den Heuvel, L, Feuillie, C, Verchere, J, Berbon, M, Arotcarena, M, Retailleau, A, Bezard, E, Laferriere, F, Loquet, A, Bousset, L, Baron, T, Lofrumento, D.D, De Giorgi, F, Stahlberg, H, Ichas, F. | | Deposit date: | 2024-03-28 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Multiple System Atrophy: Insights from aSyn Fibril Structure
To Be Published
|
|
8XBF
 
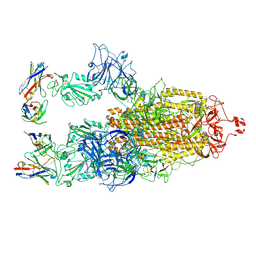 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, O5C2, heavy chain, ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8EEG
 
 | |
8EEM
 
 | |
5KWD
 
 | |
8EEH
 
 | |
8EEO
 
 | |
8EEF
 
 | |
8EEN
 
 | |
8EEI
 
 | |
8EEJ
 
 | |
8EEK
 
 | |
8EEL
 
 | |
