4APN
 
 | | Structure of TR from Leishmania infantum in complex with a diarylpirrole-based inhibitor | | Descriptor: | 4-[[1-(4-ethylphenyl)-2-methyl-5-(4-methylsulfanylphenyl)pyrrol-3-yl]methyl]thiomorpholine, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G, Biava, M. | | Deposit date: | 2012-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Leishmania Infantum Trypanothione Reductase by Azole-Based Compounds: A Comparative Analysis with its Physiological Substrate by X-Ray Crystallography.
Chemmedchem, 8, 2013
|
|
7PP0
 
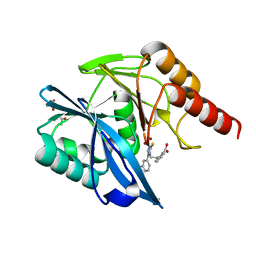 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
3RTW
 
 | |
6X8R
 
 | |
3T3I
 
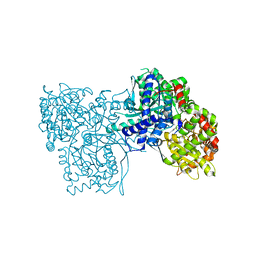 | | Glycogen Phosphorylase b in complex with GlcCF3U | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-(trifluoromethyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3G
 
 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3E
 
 | | Glycogen phosphorylase b in complex with GlcClU | | Descriptor: | 5-chloro-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
6FDK
 
 | |
3T3H
 
 | | Glycogen Phosphorylase b in complex with GlcIU | | Descriptor: | 1-(beta-D-glucopyranosyl)-5-iodopyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3T3D
 
 | | Glycogen phosphorylase b in complex with GlcU | | Descriptor: | 1-beta-D-glucopyranosylpyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
3UTU
 
 | | High affinity inhibitor of human thrombin | | Descriptor: | (2S)-N-[(4-carbamimidoylphenyl)methyl]-1-[(2S)-2-[(3-chloro-4-methoxybenzene)sulfonamido]-3-{[(4-cyanophenyl)methyl]carbamoyl}propanoyl]pyrrolidine-2-carboxamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Steinmetzer, T, Heine, A, Klebe, G. | | Deposit date: | 2011-11-26 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Beyond heparinization: design of highly potent thrombin inhibitors suitable for surface coupling
Chemmedchem, 7, 2012
|
|
6FSO
 
 | | Crystal Structure of TGT in complex with methyl({[5-(pyridin-3-yloxy)furan-2-yl]methyl})amine | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Hassaan, E, Heine, A, Klebe, G. | | Deposit date: | 2018-02-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Fragments as Novel Starting Points for tRNA-Guanine Transglycosylase Inhibitors Found by Alternative Screening Strategies.
Chemmedchem, 15, 2020
|
|
4ZQK
 
 | | Structure of the complex of human programmed death-1 (PD-1) and its ligand PD-L1. | | Descriptor: | Programmed cell death 1 ligand 1, Programmed cell death protein 1, SODIUM ION | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-05-10 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
6WS5
 
 | | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | | Descriptor: | 3-[(Z)-(2,3-difluorophenyl)diazenyl]pyridine-2,6-diamine, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | McPherson, K.S, Korzhnev, D.M. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.472 Å) | | Cite: | Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis.
Chemmedchem, 16, 2021
|
|
3RTF
 
 | |
8EGX
 
 | | Complex of Fat4(EC1-4) bound to Dchs1(EC1-4) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Medina, E, Luca, V.C. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.688 Å) | | Cite: | Structure of the planar cell polarity cadherins Fat4 and Dachsous1.
Nat Commun, 14, 2023
|
|
8EGW
 
 | | Complex of Fat4(EC1-4) bound to Dchs1(EC1-3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Medina, E, Luca, V.C. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the planar cell polarity cadherins Fat4 and Dachsous1.
Nat Commun, 14, 2023
|
|
3RT6
 
 | |
3RT8
 
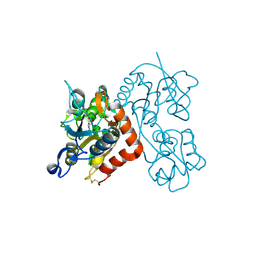 | |
5JAB
 
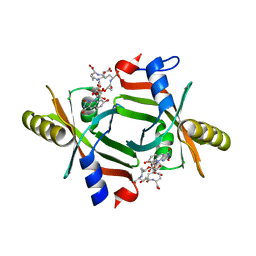 | |
1AZW
 
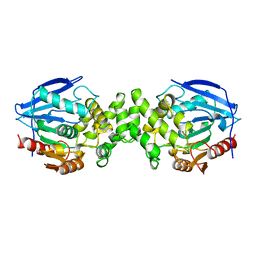 | | PROLINE IMINOPEPTIDASE FROM XANTHOMONAS CAMPESTRIS PV. CITRI | | Descriptor: | PROLINE IMINOPEPTIDASE | | Authors: | Medrano, F.J, Alonso, J, Garcia, J.L, Romero, A, Bode, W, Gomis-Ruth, F.X. | | Deposit date: | 1997-11-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of proline iminopeptidase from Xanthomonas campestris pv. citri: a prototype for the prolyl oligopeptidase family.
EMBO J., 17, 1998
|
|
4XAT
 
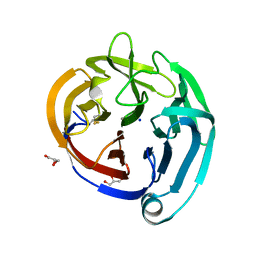 | | Crystal structure of the olfactomedin domain from noelin/pancortin/olfactomedin-1 | | Descriptor: | CALCIUM ION, GLYCEROL, Noelin, ... | | Authors: | Hill, S.E, Nguyen, E, Lieberman, R.L. | | Deposit date: | 2014-12-15 | | Release date: | 2015-07-15 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Molecular Details of Olfactomedin Domains Provide Pathway to Structure-Function Studies.
Plos One, 10, 2015
|
|
2ZXZ
 
 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-inden-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
2ZY0
 
 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-1,3-benzodisilol-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
8DFK
 
 | |
