7BMB
 
 | | Notum PPOH complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(2-prop-2-ynoxyphenyl)hexanoic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2021-01-19 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Notum Inhibitor PPOH
To Be Published
|
|
7V6M
 
 | |
7DMW
 
 | | Crystal structure of CcpC regulatory domain in complex with citrate from Bacillus amyloliquefaciens | | Descriptor: | CITRATE ANION, CcpC | | Authors: | Chen, J, Wang, L, Shang, F, Liu, W, Chen, Y, Lan, J, Bu, T, Bai, X, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Functional and structural analysis of catabolite control protein C that responds to citrate.
Sci Rep, 11, 2021
|
|
5Y8T
 
 | | Crystal structure of Bacillus subtilis PadR in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Transcriptional regulator | | Authors: | Park, S.C, Kwak, Y.M, Song, W.S, Hong, M, Yoon, S.I. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of effector and operator recognition by the phenolic acid-responsive transcriptional regulator PadR.
Nucleic Acids Res., 45, 2017
|
|
5Z5H
 
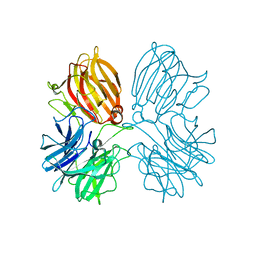 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
7WXX
 
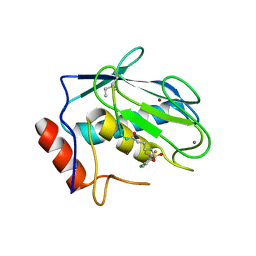 | | Crystal structure of human MMP-7 in complex with inhibitor | | Descriptor: | CALCIUM ION, Matrilysin, Peptide Inhibitor, ... | | Authors: | Kamitani, M, Oka, Y, Tabuse, H. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of Highly Potent and Selective Matrix Metalloproteinase-7 Inhibitors by Hybridizing the S1' Subsite Binder with Short Peptides.
J.Med.Chem., 65, 2022
|
|
7VLB
 
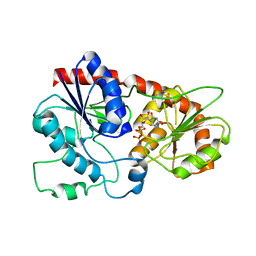 | |
5Z5F
 
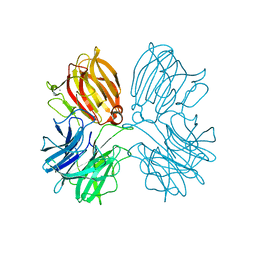 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose | | Descriptor: | Beta-xylosidase, CALCIUM ION, beta-L-arabinofuranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
7W9Z
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nitrate | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITRATE ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
5ZOB
 
 | |
7W9X
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with nickel | | Descriptor: | Iron-containing alcohol dehydrogenase, NICKEL (II) ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
7W9Y
 
 | | Crystal structure of Bacillus subtilis YugJ in complex with NADP and nickel | | Descriptor: | Iron-containing alcohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICKEL (II) ION | | Authors: | Cho, H.Y, Nam, M.S, Hong, H.J, Song, W.S, Yoon, S.I. | | Deposit date: | 2021-12-11 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and Biochemical Analysis of the Furan Aldehyde Reductase YugJ from Bacillus subtilis.
Int J Mol Sci, 23, 2022
|
|
5Z5D
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 | | Descriptor: | Beta-xylosidase, CALCIUM ION, GLYCEROL | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5I
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
6OPQ
 
 | | CD4- and 17-bound HIV-1 Env B41 SOSIP frozen with LMNG | | Descriptor: | 17b Fab heavy chain, 17b Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2019-04-25 | | Release date: | 2020-10-21 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A Strain-Specific Inhibitor of Receptor-Bound HIV-1 Targets a Pocket near the Fusion Peptide.
Cell Rep, 33, 2020
|
|
3DGA
 
 | | Wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with RJF01302, NADPH, and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, N-[2-chloro-5-(trifluoromethyl)phenyl]imidodicarbonimidic diamide, ... | | Authors: | Dasgupta, T, Chitnumsub, P, Maneeruttanarungroj, C, Kamchonwongpaisan, S, Nichols, S, Lyons, T.M, Tirado-Rives, J, Jorgensen, W.L, Yuthavong, Y, Anderson, K.S. | | Deposit date: | 2008-06-13 | | Release date: | 2009-01-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploiting structural analysis, in silico screening, and serendipity to identify novel inhibitors of drug-resistant falciparum malaria.
Acs Chem.Biol., 4, 2009
|
|
5PB1
 
 | | Crystal Structure of Factor VIIa in complex with benzenecarboximidamide | | Descriptor: | BENZAMIDINE, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Stihle, M, Mayweg, A, Roever, S, Rudolph, M.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-06-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Factor VIIa complex
To be published
|
|
5XWZ
 
 | | Crystal structure of a lactonase from Cladophialophora bantiana | | Descriptor: | GLYCEROL, MALONATE ION, SODIUM ION, ... | | Authors: | Zheng, Y.Y, Liu, W.T, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-06-30 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization and crystal structure of a novel zearalenone hydrolase from Cladophialophora bantiana
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6L7M
 
 | |
1IAK
 
 | |
5XMW
 
 | | Selenomethionine-derivated ZHD | | Descriptor: | Zearalenone lactonase | | Authors: | Hu, X.J. | | Deposit date: | 2017-05-16 | | Release date: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4W51
 
 | | T4 Lysozyme L99A with No Ligand Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W53
 
 | | T4 Lysozyme L99A with Toluene Bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endolysin, TOLUENE | | Authors: | Merski, M, Shoichet, B.K, Eidam, O, Fischer, M. | | Deposit date: | 2014-08-16 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Homologous ligands accommodated by discrete conformations of a buried cavity.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2XJJ
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(6-HYDROXY-3,3-DIMETHYL-1,2-DIHYDROINDOL-5-YL)METHANONE, GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XK2
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK PROTEIN HSP 90-ALPHA, MAGNESIUM ION | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fragment-Based Drug Discovery Applied to Hsp90. Discovery of Two Lead Series with High Ligand Efficiency.
J.Med.Chem., 53, 2010
|
|
