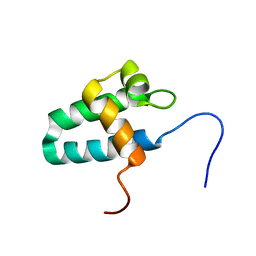
4BH9
 
 | |
4BD3
 
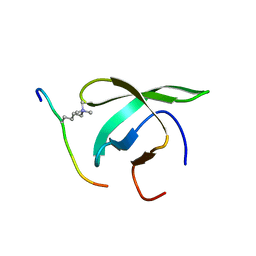 | |
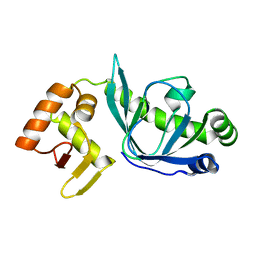
4BF8
 
 | | Fpr4 PPI domain | | Descriptor: | FPR4 | | Authors: | Monneau, Y, Mackereth, C. | | Deposit date: | 2013-03-15 | | Release date: | 2013-07-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and Activity of the Peptidyl-Prolyl Isomerase Domain from the Histone Chaperone Fpr4 Towards Histone H3 Proline Isomerization
J.Biol.Chem., 288, 2013
|
|
1NZP
 
 | | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda | | Descriptor: | DNA polymerase lambda | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Bebenek, K, Garcia-Diaz, M, Blanco, L, Kunkel, T.A, London, R.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-08-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Lyase Domain of Human DNA Polymerase Lambda
Biochemistry, 42, 2003
|
|
1C95
 
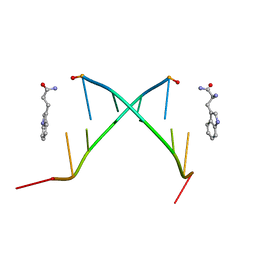 | |
1PSM
 
 | |
6GW7
 
 | | The CTD of HpDprA, a DNA binding Winged Helix domain which do not bind dsDNA | | Descriptor: | DNA protecting protein DprA | | Authors: | Lisboa, J, Celma, L, Sanchez, D, Marquis, M, Andreani, J, Guerois, R, Ochsenbein, F, Durand, D, Marsin, S, Cuniasse, P, Radicella, J.P, Quevillon-Cheruel, S. | | Deposit date: | 2018-06-22 | | Release date: | 2019-04-24 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of HpDprA is a DNA-binding winged helix domain that does not bind double-stranded DNA.
Febs J., 286, 2019
|
|
1OSW
 
 | | The Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*CP*GP*CP*UP*AP*CP*GP*GP*CP*GP*AP*GP*GP*CP*UP*CP*CP*A)-3' | | Authors: | Yuan, Y, Kerwood, D.J, Paoletti, A.C, Shubsda, M.F, Borer, P.N. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Biochemistry, 42, 2003
|
|
6J4I
 
 | |
6JIC
 
 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | Descriptor: | WCI | | Authors: | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
1CE4
 
 | |
1CG7
 
 | | HMG PROTEIN NHP6A FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | PROTEIN (NON HISTONE PROTEIN 6 A) | | Authors: | Allain, F.H.T, Yen, Y.M, Masse, J.E, Schultze, P, Dieckmann, T, Johnson, R.C, Feigon, J. | | Deposit date: | 1999-03-27 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG protein NHP6A and its interaction with DNA reveals the structural determinants for non-sequence-specific binding.
EMBO J., 18, 1999
|
|
1S79
 
 | | Solution structure of the central RRM of human La protein | | Descriptor: | Lupus La protein | | Authors: | Alfano, C, Sanfelice, D, Babon, J, Kelly, G, Jacks, A, Curry, S, Conte, M.R. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein.
Nat.Struct.Mol.Biol., 11, 2004
|
|
5XDI
 
 | |
1E68
 
 | | Solution structure of bacteriocin AS-48 | | Descriptor: | AS-48 PROTEIN | | Authors: | Gonzalez, C, Langdon, G, Bruix, M, Galvez, A, Valdivia, E, Maqueda, M, Rico, M. | | Deposit date: | 2000-08-09 | | Release date: | 2000-10-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Bacteriocin as-48, a Microbial Cyclic Polypeptide Structurally and Functionally Related to Mammalian Nk-Lysin
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5XBD
 
 | |
2RPW
 
 | | Structure of a peptide derived from H+-V-ATPase subunit a | | Descriptor: | 25 meric peptide from V-type proton ATPase subunit a, vacuolar isoform | | Authors: | Vermeer, L.S, Reat, V, Hemminga, M.A, Milon, A. | | Deposit date: | 2008-11-08 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural properties of a peptide derived from H(+)-V-ATPase subunit a
Biochim.Biophys.Acta, 1788, 2009
|
|
1ENW
 
 | |
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
5X4F
 
 | |
5XK4
 
 | | Retracted state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Dong, X, Gong, Z, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XK5
 
 | | Relaxed state of S65-phosphorylated ubiquitin | | Descriptor: | Polyubiquitin-B | | Authors: | Xu, D, Zhou, G, Qin, L.Y, Ran, M.L, Zhang, C.L, Liu, K, Liu, Z, Zhang, W.P, Tang, C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin S65 phosphorylation engenders a pH-sensitive conformational switch
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XIV
 
 | | Beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba | | Descriptor: | beta-ginkgotide, beta-gB1 | | Authors: | Wong, K.H, Tan, W.L, Xiao, T, Tam, J.P. | | Deposit date: | 2017-04-27 | | Release date: | 2017-05-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | beta-Ginkgotides: Hyperdisulfide-constrained peptides from Ginkgo biloba.
Sci Rep, 7, 2017
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
1F3C
 
 | |
