8WQI
 
 | |
8WCQ
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in intermediate state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
1YOW
 
 | | human Steroidogenic Factor 1 LBD with bound Co-factor Peptide | | Descriptor: | PHOSPHATIDYL ETHANOL, Steroidogenic factor 1, TIF2 peptide | | Authors: | Krylova, I.N, Sablin, E.P, Xu, R.X, Waitt, G.M, Juzumiene, D, Williams, J.D, Ingraham, H.A, Willson, T.M, Williams, S.P, Montana, V, Madauss, K.P, Moore, J, Bynum, J.M, Lebedeva, L, MacKay, J.A, Suzawa, M, Guy, R.K, Thornton, J.W. | | Deposit date: | 2005-01-28 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Cell(Cambridge,Mass.), 120, 2005
|
|
9BIY
 
 | |
8WCR
 
 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in open state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-09-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
2I0D
 
 | | Crystal structure of AD-81 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
5OBT
 
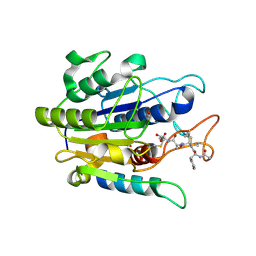 | |
8JDW
 
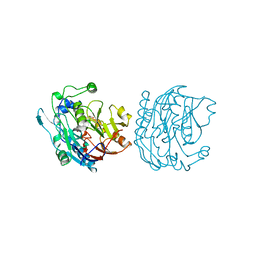 | |
2I0A
 
 | | Crystal Structure of KB-19 complexed with wild type HIV-1 protease | | Descriptor: | (5S)-3-(4-ACETYLPHENYL)-N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL]-2-OXO-1,3-OXAZOLIDINE-5-CARBOXAMIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A, Ali, A, Reddy, K.K, Cao, H, Anjum, S.G, Rana, T.M. | | Deposit date: | 2006-08-10 | | Release date: | 2006-12-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of HIV-1 Protease Inhibitors with Picomolar Affinities Incorporating N-Aryl-oxazolidinone-5-carboxamides as Novel P2 Ligands.
J.Med.Chem., 49, 2006
|
|
6FLR
 
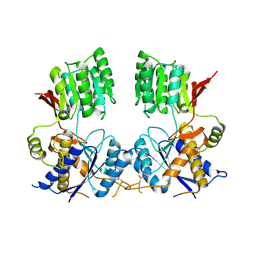 | | Super-open structure of the AMPAR GluA3 N-terminal domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 3 | | Authors: | Garcia-Nafria, J. | | Deposit date: | 2018-01-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Druggability Simulations and X-Ray Crystallography Reveal a Ligand-Binding Site in the GluA3 AMPA Receptor N-Terminal Domain.
Structure, 27, 2019
|
|
3FO4
 
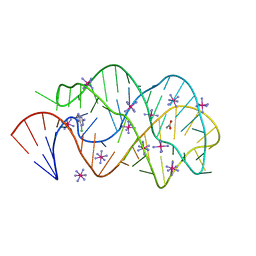 | |
2OM5
 
 | |
1JK4
 
 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
2IRX
 
 | |
1H4N
 
 | | H94N CARBONIC ANHYDRASE II COMPLEXED WITH TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
7QU7
 
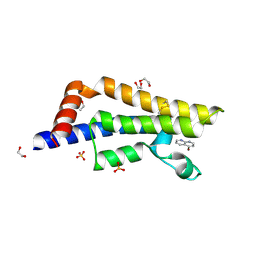 | | ATAD2 in complex with FragLite16 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,8-naphthyridine, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QWO
 
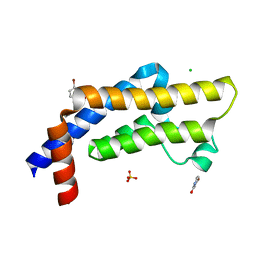 | | ATAD2 in complex with FragLite6 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1~{H}-pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-25 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QX1
 
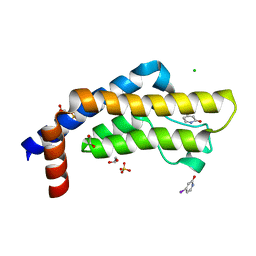 | | ATAD2 in complex with FragLite7 | | Descriptor: | 1,2-ETHANEDIOL, 4-iodanyl-3~{H}-pyridin-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QXT
 
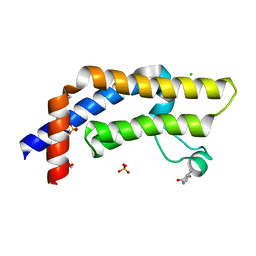 | | ATAD2 in complex with FragLite10 | | Descriptor: | 1,2-ETHANEDIOL, 6-bromo-1,3-dihydro-2H-indol-2-one, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QYL
 
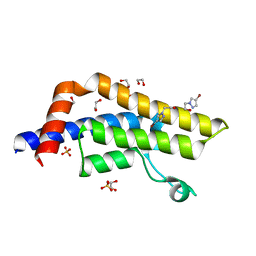 | | ATAD2 in complex with FragLite23 | | Descriptor: | 1,2-ETHANEDIOL, 2-(4-bromo-1H-pyrazol-1-yl)ethan-1-ol, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7QUK
 
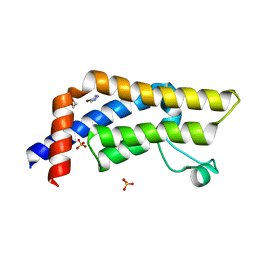 | | ATAD2 in complex with FragLite1 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromo-1H-pyrazole, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-18 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
1YKL
 
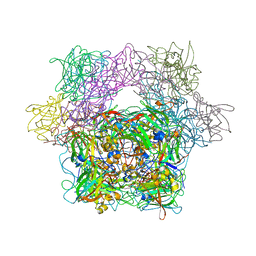 | | Protocatechuate 3,4-Dioxygenase Y408C mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKK
 
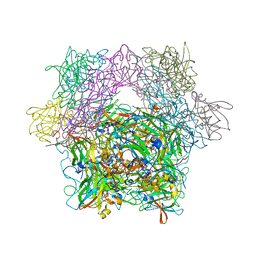 | | Protocatechuate 3,4-Dioxygenase Y408C Mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKM
 
 | | Protocatechuate 3,4-Dioxygenase Y408E mutant | | Descriptor: | FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, Protocatechuate 3,4-dioxygenase beta chain | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
1YKN
 
 | | Protocatechuate 3,4-dioxygenase Y408E mutant bound to DHB | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (III) ION, Protocatechuate 3,4-dioxygenase alpha chain, ... | | Authors: | Brown, C.K, Ohlendorf, D.H. | | Deposit date: | 2005-01-18 | | Release date: | 2005-08-16 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Roles of the equatorial tyrosyl iron ligand of protocatechuate 3,4-dioxygenase in catalysis
Biochemistry, 44, 2005
|
|
