1FC0
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE COMPLEXED WITH N-ACETYL-BETA-D-GLUCOPYRANOSYLAMINE | | Descriptor: | GLYCOGEN PHOSPHORYLASE, LIVER FORM, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, LeMotte, P.K, Fennell, K.F, Mansour, M.M, Danley, D.E, Hynes, T.R, Schulte, G.K, Wasilko, D.J, Pandit, J. | | Deposit date: | 2000-07-17 | | Release date: | 2000-08-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation of human liver glycogen phosphorylase by alteration of the secondary structure and packing of the catalytic core.
Mol.Cell, 6, 2000
|
|
1FC1
 
 | |
1FC2
 
 | |
1FC3
 
 | |
1FC4
 
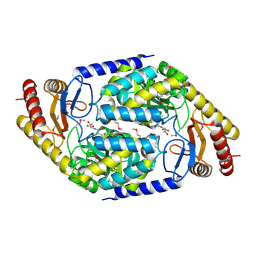 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1FC5
 
 | |
1FC6
 
 | | PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE | | Descriptor: | PHOTOSYSTEM II D1 PROTEASE | | Authors: | Liao, D.I, Qian, J, Chisholm, D.A, Jordan, D.B, Diner, B.A. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-18 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the photosystem II D1 C-terminal processing protease.
Nat.Struct.Biol., 7, 2000
|
|
1FC7
 
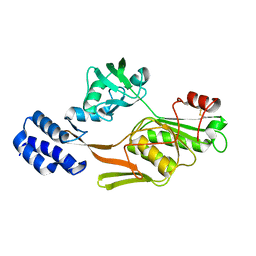 | | PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE | | Descriptor: | PHOTOSYSTEM II D1 PROTEASE | | Authors: | Liao, D.I, Qian, J, Chisholm, D.A, Jordan, D.B, Diner, B.A. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-18 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the photosystem II D1 C-terminal processing protease.
Nat.Struct.Biol., 7, 2000
|
|
1FC8
 
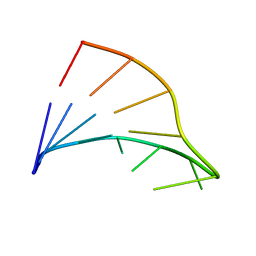 | |
1FC9
 
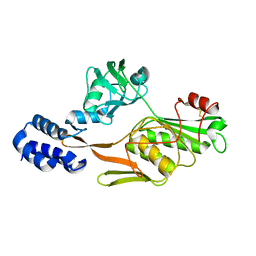 | | PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE | | Descriptor: | PHOTOSYSTEM II D1 PROTEASE | | Authors: | Liao, D.I, Qian, J, Chisholm, D.A, Jordan, D.B, Diner, B.A. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-18 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the photosystem II D1 C-terminal processing protease.
Nat.Struct.Biol., 7, 2000
|
|
1FCA
 
 | |
1FCB
 
 | |
1FCC
 
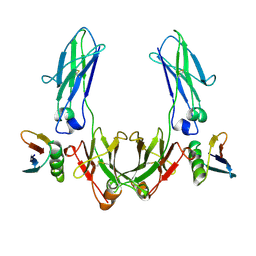 | | CRYSTAL STRUCTURE OF THE C2 FRAGMENT OF STREPTOCOCCAL PROTEIN G IN COMPLEX WITH THE FC DOMAIN OF HUMAN IGG | | Descriptor: | IGG1 MO61 FC, STREPTOCOCCAL PROTEIN G (C2 FRAGMENT) | | Authors: | Sauer-Eriksson, A.E, Kleywegt, G.J, Uhlen, M, Jones, T.A. | | Deposit date: | 1995-01-17 | | Release date: | 1995-04-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG.
Structure, 3, 1995
|
|
1FCD
 
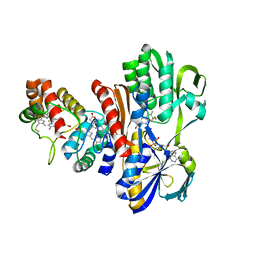 | | THE STRUCTURE OF FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE FROM A PURPLE PHOTOTROPHIC BACTERIUM CHROMATIUM VINOSUM AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (CYTOCHROME SUBUNIT), FLAVOCYTOCHROME C SULFIDE DEHYDROGENASE (FLAVIN-BINDING SUBUNIT), ... | | Authors: | Chen, Z.W, Koh, M, Van Driessche, G, Van Beeumen, J.J, Bartsch, R.G, Meyer, T.E, Cusanovich, M.A, Mathews, F.S. | | Deposit date: | 1994-08-18 | | Release date: | 1994-11-01 | | Last modified: | 2021-03-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of flavocytochrome c sulfide dehydrogenase from a purple phototrophic bacterium.
Science, 266, 1994
|
|
1FCE
 
 | | PROCESSIVE ENDOCELLULASE CELF OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, CELLULASE CELF, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside | | Authors: | Parsiegla, G, Juy, M, Haser, R. | | Deposit date: | 1998-07-06 | | Release date: | 1999-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the processive endocellulase CelF of Clostridium cellulolyticum in complex with a thiooligosaccharide inhibitor at 2.0 A resolution.
EMBO J., 17, 1998
|
|
1FCF
 
 | | PHOTOSYSTEM II D1 C-TERMINAL PROCESSING PROTEASE | | Descriptor: | PHOTOSYSTEM II D1 PROTEASE, SULFATE ION | | Authors: | Liao, D.I, Qian, J, Chisholm, D.A, Jordan, D.B, Diner, B.A. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-18 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the photosystem II D1 C-terminal processing protease.
Nat.Struct.Biol., 7, 2000
|
|
1FCG
 
 | | ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA | | Descriptor: | PROTEIN (FC RECEPTOR FC(GAMMA)RIIA) | | Authors: | Maxwell, K.F, Powell, M.S, Garrett, T.P, Hogarth, P.M. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa.
Nat.Struct.Biol., 6, 1999
|
|
1FCH
 
 | | CRYSTAL STRUCTURE OF THE PTS1 COMPLEXED TO THE TPR REGION OF HUMAN PEX5 | | Descriptor: | PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR, PTS1-CONTAINING PEPTIDE | | Authors: | Gatto Jr, G.J, Geisbrecht, B.V, Gould, S.J, Berg, J.M. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5.
Nat.Struct.Biol., 7, 2000
|
|
1FCJ
 
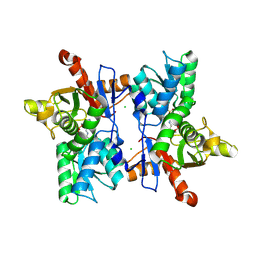 | | CRYSTAL STRUCTURE OF OASS COMPLEXED WITH CHLORIDE AND SULFATE | | Descriptor: | CHLORIDE ION, O-ACETYLSERINE SULFHYDRYLASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Burkhard, P, Tai, C, Jansonius, J.N, Cook, P.F. | | Deposit date: | 2000-07-18 | | Release date: | 2000-10-18 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of an allosteric anion-binding site on O-acetylserine sulfhydrylase: structure of the enzyme with chloride bound.
J.Mol.Biol., 303, 2000
|
|
1FCK
 
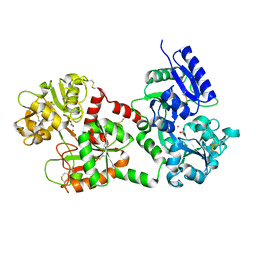 | | STRUCTURE OF DICERIC HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, CERIUM (III) ION, LACTOFERRIN | | Authors: | Baker, H.M, Baker, C.J, Smith, C.A, Baker, E.N. | | Deposit date: | 2000-07-18 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metal substitution in transferrins: specific binding of cerium(IV) revealed by the crystal structure of cerium-substituted human lactoferrin.
J.Biol.Inorg.Chem., 5, 2000
|
|
1FCL
 
 | |
1FCM
 
 | |
1FCN
 
 | |
1FCO
 
 | | CRYSTAL STRUCTURE OF THE E. COLI AMPC BETA-LACTAMASE COVALENTLY ACYLATED WITH THE INHIBITORY BETA-LACTAM, MOXALACTAM | | Descriptor: | (2R)-2-[(1R)-1-{[(2S)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, BETA-LACTAMASE | | Authors: | Patera, A, Blaszczak, L.C, Shoichet, B.K. | | Deposit date: | 2000-07-19 | | Release date: | 2000-12-04 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Substrate and Inhibitor Complexes with AmpC -Lactamase: Possible Implications for Substrate-Assisted Catalysis
J.Am.Chem.Soc., 122, 2000
|
|
1FCP
 
 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-14 | | Release date: | 1999-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
