1F5X
 
 | | NMR STRUCTURE OF THE Y174 AUTOINHIBITED DBL HOMOLOGY DOMAIN | | Descriptor: | RHO-GEF VAV | | Authors: | Aghazadeh, B, Rosen, M.K, Lowry, W.E, Huang, X.Y. | | Deposit date: | 2000-06-18 | | Release date: | 2000-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation.
Cell(Cambridge,Mass.), 102, 2000
|
|
1F5Y
 
 | | NMR STRUCTURE OF A CONCATEMER OF THE FIRST AND SECOND LIGAND-BINDING MODULES OF THE HUMAN LDL RECEPTOR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | Kurniawan, N.D, Atkins, A.R, Brereton, I.M, Kroon, P.A, Smith, R. | | Deposit date: | 2000-06-18 | | Release date: | 2000-08-30 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a concatemer of the first and second ligand-binding modules of the human low-density lipoprotein receptor.
Protein Sci., 9, 2000
|
|
1F5Z
 
 | |
1F60
 
 | | CRYSTAL STRUCTURE OF THE YEAST ELONGATION FACTOR COMPLEX EEF1A:EEF1BA | | Descriptor: | ELONGATION FACTOR EEF1A, ELONGATION FACTOR EEF1BA | | Authors: | Andersen, G.R, Pedersen, L, Valente, L, Kinzy, T.G, Nyborg, J. | | Deposit date: | 2000-06-19 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Balpha.
Mol.Cell, 6, 2000
|
|
1F61
 
 | | CRYSTAL STRUCTURE OF ISOCITRATE LYASE FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ISOCITRATE LYASE, MAGNESIUM ION | | Authors: | Sharma, V, Sharma, S, Hoener zu Bentrup, K.H, McKinney, J.D, Russell, D.G, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-06-19 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis.
Nat.Struct.Biol., 7, 2000
|
|
1F62
 
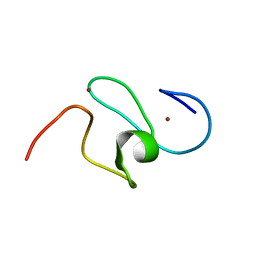 | | WSTF-PHD | | Descriptor: | TRANSCRIPTION FACTOR WSTF, ZINC ION | | Authors: | Pascual, J, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-19 | | Release date: | 2000-12-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor.
J.Mol.Biol., 304, 2000
|
|
1F63
 
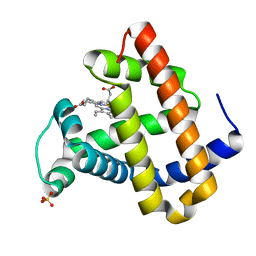 | | CRYSTAL STRUCTURE OF DEOXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
1F65
 
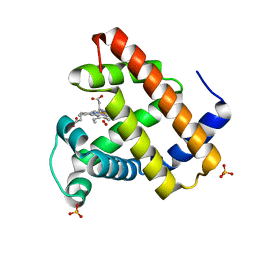 | | CRYSTAL STRUCTURE OF OXY SPERM WHALE MYOGLOBIN MUTANT Y(B10)Q(E7)R(E10) | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brunori, M, Cutruzzola, F, Savino, C, Travaglini-Allocatelli, C, Vallone, B, Gibson, Q.H. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural dynamics of ligand diffusion in the protein matrix: A study on a new myoglobin mutant Y(B10) Q(E7) R(E10).
Biophys.J., 76, 1999
|
|
1F66
 
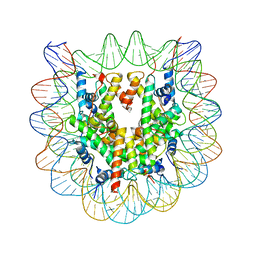 | | 2.6 A CRYSTAL STRUCTURE OF A NUCLEOSOME CORE PARTICLE CONTAINING THE VARIANT HISTONE H2A.Z | | Descriptor: | HISTONE H2A.Z, HISTONE H2B, HISTONE H3, ... | | Authors: | Suto, R.K, Clarkson, M.J, Tremethick, D.J, Luger, K. | | Deposit date: | 2000-06-20 | | Release date: | 2000-11-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a nucleosome core particle containing the variant histone H2A.Z.
Nat.Struct.Biol., 7, 2000
|
|
1F68
 
 | |
1F69
 
 | |
1F6A
 
 | | Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR ALPHA-SUBUNIT, ... | | Authors: | Garman, S.C, Wurzburg, B.A, Tarchevskaya, S.S, Kinet, J.P, Jardetzky, T.S. | | Deposit date: | 2000-06-20 | | Release date: | 2000-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc (epsilon) RI (alpha).
Nature, 406, 2000
|
|
1F6B
 
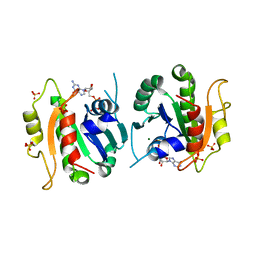 | | CRYSTAL STRUCTURE OF SAR1-GDP COMPLEX | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SAR1, ... | | Authors: | Huang, M, Wilson, I.A, Balch, W.E. | | Deposit date: | 2000-06-21 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH2 terminus in ER export.
J.Cell Biol., 155, 2001
|
|
1F6C
 
 | |
1F6D
 
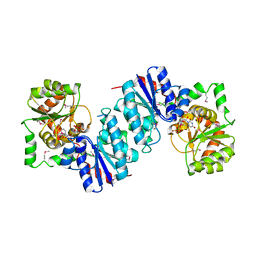 | | THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI. | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE, ... | | Authors: | Campbell, R.E, Mosimann, S.C, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases.
Biochemistry, 39, 2000
|
|
1F6E
 
 | |
1F6F
 
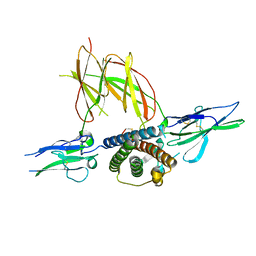 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX BETWEEN OVINE PLACENTAL LACTOGEN AND THE EXTRACELLULAR DOMAIN OF THE RAT PROLACTIN RECEPTOR | | Descriptor: | PLACENTAL LACTOGEN, PROLACTIN RECEPTOR | | Authors: | Elkins, P.A, Christinger, H.W, Sandowski, Y, Sakal, E, Gertler, A, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2000-06-21 | | Release date: | 2000-07-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ternary complex between placental lactogen and the extracellular domain of the prolactin receptor.
Nat.Struct.Biol., 7, 2000
|
|
1F6G
 
 | | POTASSIUM CHANNEL (KCSA) FULL-LENGTH FOLD | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL | | Authors: | Cortes, D.M, Perozo, E. | | Deposit date: | 2000-06-21 | | Release date: | 2001-02-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating.
J.Gen.Physiol., 117, 2001
|
|
1F6H
 
 | |
1F6I
 
 | |
1F6J
 
 | |
1F6K
 
 | | CRYSTAL STRUCTURE ANALYSIS OF N-ACETYLNEURAMINATE LYASE FROM HAEMOPHILUS INFLUENZAE: CRYSTAL FORM II | | Descriptor: | GLYCEROL, N-ACETYLNEURAMINATE LYASE, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Smith, B.J, DeGori, R, Lawrence, M.C. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Active site modulation in the N-acetylneuraminate lyase sub-family as revealed by the structure of the inhibitor-complexed Haemophilus influenzae enzyme.
J.Mol.Biol., 303, 2000
|
|
1F6L
 
 | | VARIABLE LIGHT CHAIN DIMER OF ANTI-FERRITIN ANTIBODY | | Descriptor: | ANTI-FERRITIN IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Nymalm, Y, Kravchuk, Z, Salminen, T, Chumanevich, A.A, Dubnovitsky, A.P, Kankare, J, Pentikainen, O, Lehtonen, J, Arosio, P, Martsev, S, Johnson, M.S. | | Deposit date: | 2000-06-22 | | Release date: | 2002-10-23 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antiferritin VL homodimer binds human spleen ferritin with high specificity
J.STRUCT.BIOL., 138, 2002
|
|
1F6M
 
 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THIOREDOXIN REDUCTASE, THIOREDOXIN, AND THE NADP+ ANALOG, AADP+ | | Descriptor: | 3-AMINOPYRIDINE-ADENINE DINUCLEOTIDE PHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN 1, ... | | Authors: | Lennon, B.W, Williams Jr, C.H, Ludwig, M.L. | | Deposit date: | 2000-06-22 | | Release date: | 2000-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase.
Science, 289, 2000
|
|
1F6N
 
 | | CRYSTAL STRUCTURE ANALYSIS OF THE MUTANT REACTION CENTER PRO L209-> TYR FROM THE PHOTOSYNTHETIC PURPLE BACTERIUM RHODOBACTER SPHAEROIDES | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Kuglstatter, A, Ermler, U, Michel, H, Baciou, L, Fritzsch, G. | | Deposit date: | 2000-06-22 | | Release date: | 2001-04-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure analyses of photosynthetic reaction center variants from Rhodobacter sphaeroides: structural changes induced by point mutations at position L209 modulate electron and proton transfer.
Biochemistry, 40, 2001
|
|
