5RH9
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4438424255 (Mpro-x2776) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-{4-[(1S)-1-methoxyethyl]phenyl}-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RL2
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-26 (Mpro-x3115) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(2-methoxyethyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL0
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-2 (Mpro-x3110) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5QI0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of human PARP14 Macrodomain 3 in complex with FMOPL000352a | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 14, ... | | Authors: | Schuller, M, Talon, R, Krojer, T, Brandao-Neto, J, Douangamath, A, Zhang, R, von Delft, F, Schuler, H, Kessler, B, Knapp, S, Bountra, C, Arrowsmith, C.H, Edwards, A, Elkins, J. | | Deposit date: | 2018-05-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QRH
 
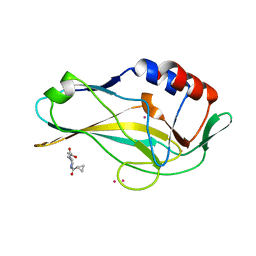 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z32327641 | | Descriptor: | CADMIUM ION, T-box transcription factor T, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QS7
 
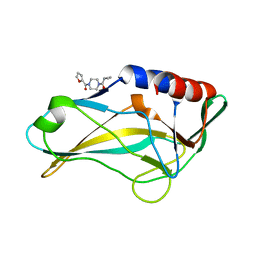 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z32327641 | | Descriptor: | T-box transcription factor T, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RH3
 
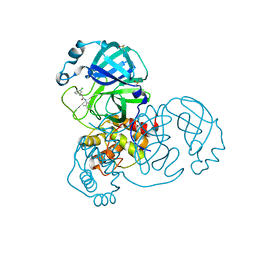 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1264525706 (Mpro-x2649) | | Descriptor: | (2R)-2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)propanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
7HLA
 
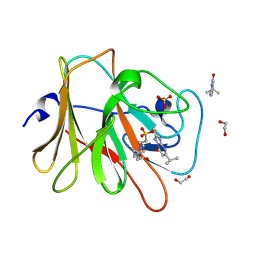 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z111634612 | | Descriptor: | (8R)-2-methyl-5-(propan-2-yl)[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HLT
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z328695024 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, N,1-dimethyl-N-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RH5
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4439011520 (Mpro-x2694) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(5-tert-butyl-1,2-oxazol-3-yl)-N-[(1R)-2-[(4-methoxy-2-methylphenyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RKW
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z27678561 | | Descriptor: | N-(6-methoxy-1,3-benzothiazol-2-yl)cyclopropanecarboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.242 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
7HNP
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z106579662 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, SULFATE ION, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
7HO9
 
 | | PanDDA analysis group deposition -- Crystal Structure of TRIM21 in complex with Z1250132788 | | Descriptor: | (1R)-N-methyl-1-(pyridin-4-yl)propan-1-amine, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM21, ... | | Authors: | Kim, Y, Marples, P, Fearon, D, von Delft, F, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-11-04 | | Release date: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RL4
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-3 (Mpro-x3124) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(methylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
7HPB
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with ASAP-0015080-001 | | Descriptor: | DIMETHYL SULFOXIDE, N-{3-chloro-5-[(1-hydroxy-2-methylpropan-2-yl)carbamoyl]phenyl}-2-methyl-3-oxo-2,3-dihydro-1,2-oxazole-5-carboxamide, Serine protease NS3, ... | | Authors: | Ni, X, Marples, P.G, Godoy, A.S, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Fairhead, M, Lithgo, R.M, Lee, A, Kenton, N, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Williams, E.P, Chandran, A.V, Walsh, M.A, Fearon, D, von Delft, F. | | Deposit date: | 2024-11-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RK8
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z53116498 | | Descriptor: | 3-(2-methyl-1H-benzimidazol-1-yl)propanamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RL1
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-27 (Mpro-x3113) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-[(3-methoxypropyl)amino]-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5RL5
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-30 (Mpro-x3359) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(ethylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5SLL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z54615640 | | Descriptor: | N-[(3R)-3-methyl-1,1-dioxo-1lambda~6~-thiolan-3-yl]cyclopropanecarboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLH
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z65532537 | | Descriptor: | (2S)-2-(2-fluorophenoxy)propanoic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z32327641 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, [4-(cyclopropanecarbonyl)piperazin-1-yl](furan-2-yl)methanone | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QSE
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z2017168803 | | Descriptor: | (2S)-2-[(3-fluoropyridin-2-yl)(methyl)amino]propan-1-ol, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QRW
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury in complex with Z1509882419 | | Descriptor: | 1-(propan-2-yl)-1H-imidazole-4-sulfonamide, CADMIUM ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Fernandez-Cid, A, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-07-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QSB
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z2856434874 | | Descriptor: | 1-{[4-(propan-2-yl)phenyl]methyl}piperidin-4-ol, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QS8
 
 | | PanDDA analysis group deposition -- Crystal Structure of human Brachyury G177D variant in complex with Z1432018343 | | Descriptor: | (2S)-2-[(5-chloro-3-fluoropyridin-2-yl)amino]propan-1-ol, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A.E, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-05-25 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
