2DCC
 
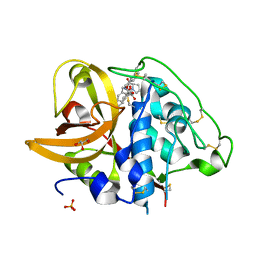 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA077 complex | | Descriptor: | BENZYL N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, CATHEPSIN B, GLYCEROL, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCD
 
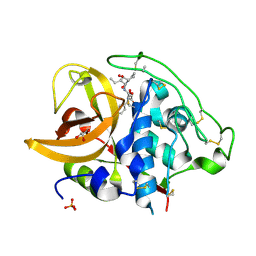 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA078 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-({(2S,3S)-3-[(BENZYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
1SNX
 
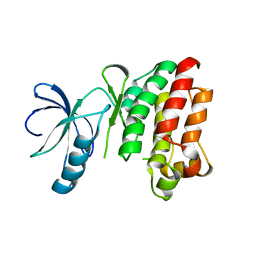 | | CRYSTAL STRUCTURE OF APO INTERLEUKIN-2 TYROSINE KINASE CATALYTIC DOMAIN | | Descriptor: | Tyrosine-protein kinase ITK/TSK | | Authors: | Brown, K, Long, J.M, Vial, S.C, Dedi, N, Dunster, N.J, Renwick, S.B, Tanner, A.J, Frantz, J.D, Fleming, M.A, Cheetham, G.M.T. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of interleukin-2 tyrosine kinase and their implications for the design of selective inhibitors.
J.Biol.Chem., 279, 2004
|
|
2DCB
 
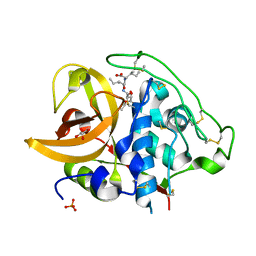 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA076 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DCA
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA075 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCYL-L-ALANINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2006-01-01 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC9
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA074Me complex | | Descriptor: | CATHEPSIN B, GLYCEROL, METHYL N-({(2S,3S)-3-[(PROPYLAMINO)CARBONYL]OXIRAN-2-YL}CARBONYL)-L-ISOLEUCYL-L-PROLINATE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
2DC8
 
 | | X-ray crystal structure analysis of bovine spleen cathepsin B-CA059 complex | | Descriptor: | CATHEPSIN B, GLYCEROL, N-{[(2S,3S)-3-(ETHOXYCARBONYL)OXIRAN-2-YL]CARBONYL}-L-ISOLEUCINE, ... | | Authors: | Watanabe, D. | | Deposit date: | 2005-12-31 | | Release date: | 2006-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Quantitative estimation of each active subsite of cathepsin B for the inhibitory activity, based on the inhibitory activitybinding mode relationship of a series of epoxysuccinyl inhibitors by X-ray crystal structure analyses of the complexes
To be Published
|
|
4GDP
 
 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
2SN3
 
 | | STRUCTURE OF SCORPION TOXIN VARIANT-3 AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, SCORPION NEUROTOXIN (VARIANT 3) | | Authors: | Zhao, B, Carson, M, Ealick, S.E, Bugg, C.E. | | Deposit date: | 1992-02-20 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of scorpion toxin variant-3 at 1.2 A resolution.
J.Mol.Biol., 227, 1992
|
|
1OSX
 
 | | Solution Structure of the Extracellular Domain of BLyS Receptor 3 (BR3) | | Descriptor: | Tumor necrosis factor receptor superfamily member 13C | | Authors: | Gordon, N.C, Pan, B, Hymowitz, S.G, Yin, J.P, Kelley, R.F, Cochran, A.G, Yan, M, Dixit, V.M, Fairbrother, W.J, Starovasnik, M.A. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site
Biochemistry, 42, 2003
|
|
1PAA
 
 | | STRUCTURE OF A HISTIDINE-X4-HISTIDINE ZINC FINGER DOMAIN: INSIGHTS INTO ADR1-UAS1 PROTEIN-DNA RECOGNITION | | Descriptor: | YEAST TRANSCRIPTION FACTOR ADR1, ZINC ION | | Authors: | Bernstein, B.E, Hoffman, R.C, Horvath, S.J, Herriott, J.R, Klevit, R.E. | | Deposit date: | 1994-07-15 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of a histidine-X4-histidine zinc finger domain: insights into ADR1-UAS1 protein-DNA recognition.
Biochemistry, 33, 1994
|
|
4HQQ
 
 | | Crystal structure of RV144-elicited antibody CH58 | | Descriptor: | CH58 Fab heavy chain, CH58 Fab light chain | | Authors: | McLellan, J.S, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
5IEM
 
 | | NMR structure of the 5'-terminal hairpin of the 7SK snRNA | | Descriptor: | 7SK snRNA | | Authors: | Bourbigot, S, Dock-Bregeon, A.C, Coutant, J, Kieffer, B, Lebars, I. | | Deposit date: | 2016-02-25 | | Release date: | 2016-11-02 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 5'-terminal hairpin of the 7SK small nuclear RNA.
RNA, 22, 2016
|
|
32C2
 
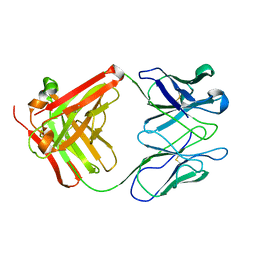 | | STRUCTURE OF AN ACTIVITY SUPPRESSING FAB FRAGMENT TO CYTOCHROME P450 AROMATASE | | Descriptor: | IGG1 ANTIBODY 32C2 | | Authors: | Sawicki, M.W, Ng, P.C, Burkhart, B, Pletnev, V, Higashiyama, T, Osawa, Y, Ghosh, D. | | Deposit date: | 1999-04-21 | | Release date: | 2000-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an activity suppressing Fab fragment to cytochrome P450 aromatase: insights into the antibody-antigen interactions.
Mol.Immunol., 36, 1999
|
|
4HQX
 
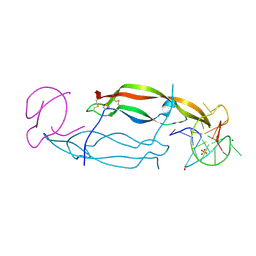 | | CRYSTAL STRUCTURE OF HUMAN PDGF-BB IN COMPLEX WITH A Modified nucleotide aptamer (SOMAmer SL4) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1NGH
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
4HPY
 
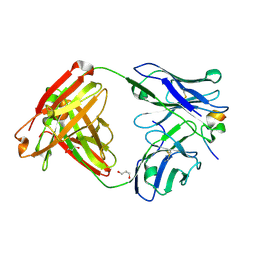 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | Descriptor: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | Authors: | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | Deposit date: | 2012-10-24 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
4HQU
 
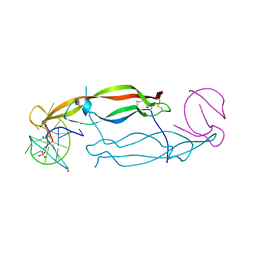 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1JXU
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 240 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-09 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXX
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 200 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JXY
 
 | | CRAMBIN MIXED SEQUENCE FORM AT 220 K. PROTEIN/WATER SUBSTATES | | Descriptor: | Crambin, ETHANOL | | Authors: | Teeter, M.M, Yamano, A, Stec, B, Mohanty, U. | | Deposit date: | 2001-09-10 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | On the nature of a glassy state of matter in a hydrated protein: Relation to protein function.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2YXY
 
 | | Crystarl structure of Hypothetical conserved protein, GK0453 | | Descriptor: | Hypothetical conserved protein, GK0453 | | Authors: | Nakamu, Y, Bessho, Y, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the hypothetical DUF1811-family protein GK0453 from Geobacillus kaustophilus HTA426
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3R3A
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant Q58A complexed with 4-hydroxybenzoic acid and CoA | | Descriptor: | 4-hydroxybenzoyl-CoA thioesterase, COENZYME A, P-HYDROXYBENZOIC ACID | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
4HB3
 
 | |
2O58
 
 | | Horse heart met manganese myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING MN, SULFATE ION | | Authors: | Richter-Addo, G.B, Zahran, Z.N, Chooback, L, Copeland, D.M, West, A.H. | | Deposit date: | 2006-12-05 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of manganese- and cobalt-substituted myoglobin in complex with NO and nitrite reveal unusual ligand conformations.
J.Inorg.Biochem., 102, 2008
|
|
