4D4T
 
 | | RSV Matrix protein | | Descriptor: | GLYCEROL, MATRIX PROTEIN, POTASSIUM ION, ... | | Authors: | Forster, A, Maertens, G, Bajorek, M. | | Deposit date: | 2014-10-31 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dimerization of Matrix Protein is Required for Budding of Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4D51
 
 | | Crystal structure of CymA from Klebsiella oxytoca | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, CYMA | | Authors: | van den Berg, B, Bhamidimarri, S.P, Kleinekathoefer, U, Winterhalter, M. | | Deposit date: | 2014-11-01 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Outer-membrane translocation of bulky small molecules by passive diffusion.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
4CV8
 
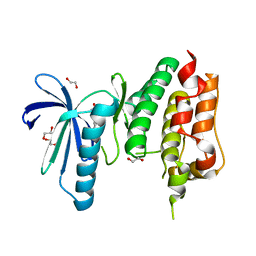 | | MPS1 kinase with 3-aminopyridin-2-one inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 3-amino-5-(1-methyl-1H-pyrazol-4-yl)pyridin-2(1H)-one, DUAL SPECIFICITY PROTEIN KINASE TTK | | Authors: | Fearon, D, Bavetsias, V, Bayliss, R, Schmitt, J, Westwood, I.M, vanMontfort, R.L.M, Jones, K. | | Deposit date: | 2014-03-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Protein Kinase Selectivity of a 3-Aminopyridin-2- One Based Fragment Library, Identification of 3-Amino-5-(Pyridin-4-Yl)Pyridin-2(1H)-One as a Novel Scaffold for Mps1 Inhibition
To be Published
|
|
4CNT
 
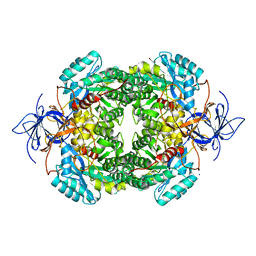 | | CRYSTAL STRUCTURE OF WT HUMAN CRMP-4 | | Descriptor: | 1,2-ETHANEDIOL, DIHYDROPYRIMIDINASE-LIKE 3, SODIUM ION | | Authors: | Ponnusamy, R, Lohkamp, B. | | Deposit date: | 2014-01-24 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of Human Crmp-4: Correction of Intensities for Lattice-Translocation Disorder
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CS1
 
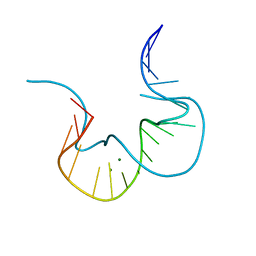 | |
4CXK
 
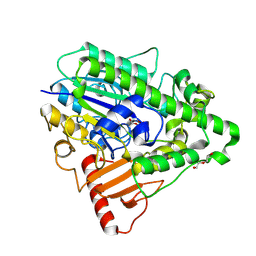 | | G9 mutant of PAS, arylsulfatase from Pseudomonas Aeruginosa | | Descriptor: | ARYLSULFATASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miton, C.M, Jonas, S, Mohammed, M.F, Fischer, G, Loo, B.v, Kintses, B, Hyvonen, M, Tokuriki, N, Hollfelder, F. | | Deposit date: | 2014-04-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Evolutionary repurposing of a sulfatase: A new Michaelis complex leads to efficient transition state charge offset.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4D3N
 
 | |
4D5U
 
 | | Structure of OmpF in I2 | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Chaptal, V, Kilburg, A, Flot, D, Wiseman, B, Aghajari, N, Jault, J.M, Falson, P. | | Deposit date: | 2014-11-07 | | Release date: | 2015-12-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Two Different Centered Monoclinic Crystals of the E. Coli Outer-Membrane Protein Ompf Originate from the Same Building Block.
Biochim.Biophys.Acta, 1858, 2016
|
|
4D73
 
 | | X-ray structure of a peroxiredoxin | | Descriptor: | 1-CYS PEROXIREDOXIN | | Authors: | Staudacher, V, Djuika, C.F, Koduka, J, Schlossarek, S, Kopp, J, Buechler, M, Lanzer, M, Deponte, M. | | Deposit date: | 2014-11-19 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Plasmodium Falciparum Antioxidant Protein Reveals a Novel Mechanism for Balancing Turnover and Inactivation of Peroxiredoxins
Free Radic.Biol.Med., 85, 2015
|
|
4D04
 
 | | Structure of the Cys65Asp mutant of phenylacetone monooxygenase: reduced state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, HEXAETHYLENE GLYCOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Brondani, P.B, Dudek, H.M, Martinoli, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2014-04-24 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Finding the Switch: Turning a Baeyer-Villiger Monooxygenase Into a Nadph Oxidase.
J.Am.Chem.Soc., 136, 2014
|
|
4D7H
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 7-(2-(3-(3-Fluorophenyl(propylamino)ethyl))quinolin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[2-[3-(3-fluorophenyl)propylamino]ethyl]quinolin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Nitric Oxide Synthase as a Target for Methicillin-Resistant Staphylococcus Aureus
Chem.Biol., 22, 2015
|
|
4D0M
 
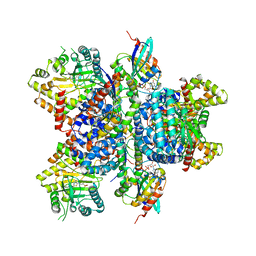 | | Phosphatidylinositol 4-kinase III beta in a complex with Rab11a-GTP- gamma-S and the Rab-binding domain of FIP3 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, ... | | Authors: | Burke, J.E, Inglis, A.J, Perisic, O, Masson, G.R, McLaughlin, S.H, Rutaganira, F, Shokat, K.M, Williams, R.L. | | Deposit date: | 2014-04-29 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structures of Pi4Kiiibeta Complexes Show Simultaneous Recruitment of Rab11 and its Effectors.
Science, 344, 2014
|
|
4CX9
 
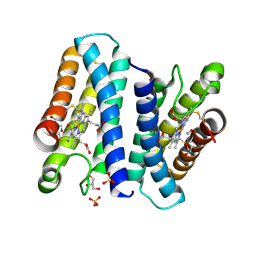 | | The 5-coordinate proximal NO complex of cytochrome c prime from Shewanella frigidimarina | | Descriptor: | CYTOCHROME C, CLASS II, GLYCEROL, ... | | Authors: | Manole, A.A, Kekilli, D, Dobbin, P.S, Hough, M.A. | | Deposit date: | 2014-04-04 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Conformational Control of the Binding of Diatomic Gases to Cytochrome C'.
J.Biol.Inorg.Chem., 20, 2015
|
|
4CY7
 
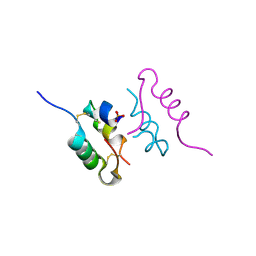 | | Crystal structure of human insulin analogue (NMe-AlaB8)-insulin crystal form II | | Descriptor: | ACETATE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Kosinova, L, Veverka, V, Novotna, P, Collinsova, M, Urbanova, M, Jiracek, J, Moody, N.R, Turkenburg, J.P, Brzozowski, A.M, Zakova, L. | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Insight Into Structural and Biological Relevance of the T/R Transition of the B-Chain N-Terminus in Human Insulin.
Biochemistry, 53, 2014
|
|
4D3I
 
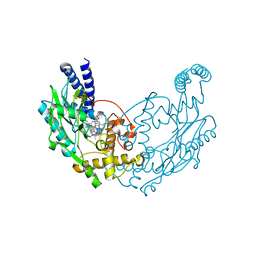 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-((5-(aminomethyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(aminomethyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), GLYCEROL, N-PROPANOL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3U
 
 | |
4D7J
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6-(4-(((3-Fluorophenethyl)amino)methyl)phenyl)-4-methylpyridin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[4-({[2-(3-fluorophenyl)ethyl]amino}methyl)phenyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Nitric Oxide Synthase as a Target for Methicillin-Resistant Staphylococcus Aureus.
Chem.Biol., 22, 2015
|
|
4D7I
 
 | | Structure of Bacillus subtilis nitric oxide synthase I218V in complex with 6-(4-(((3-Fluorophenethyl)amino)methyl)phenyl)-4-methylpyridin-2- amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[4-({[2-(3-fluorophenyl)ethyl]amino}methyl)phenyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-11-25 | | Release date: | 2015-07-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nitric Oxide Synthase as a Target for Methicillin-Resistant Staphylococcus Aureus
Chem.Biol., 22, 2015
|
|
4D1E
 
 | | THE CRYSTAL STRUCTURE OF HUMAN MUSCLE ALPHA-ACTININ-2 | | Descriptor: | ALPHA-ACTININ-2 | | Authors: | Pinotsis, N, Salmazo, A, Sjoeblom, B, Gkougkoulia, E, Djinovic-Carugo, K. | | Deposit date: | 2014-05-01 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure and Regulation of Human Muscle Alpha-Actinin
Cell(Cambridge,Mass.), 159, 2014
|
|
4D79
 
 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with ATP at 1.768 Angstroem resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
5OW8
 
 | | Indole-2 carboxamides as selective secreted phospholipase A2 type X (sPLA2-X) inhibitors | | Descriptor: | 1-[3-(trifluoromethyl)phenyl]indole-2-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sandmark, J.S, Roth, R.G, Knerr, L, Bodin, C, Pettersen, D. | | Deposit date: | 2017-08-31 | | Release date: | 2018-08-01 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of a Series of Indole-2 Carboxamides as Selective Secreted Phospholipase A2Type X (sPLA2-X) Inhibitors.
Acs Med.Chem.Lett., 9, 2018
|
|
4D3T
 
 | |
4D4A
 
 | | Structure of the catalytic domain (BcGH76) of the Bacillus circulans GH76 alpha mannanase, Aman6. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D7A
 
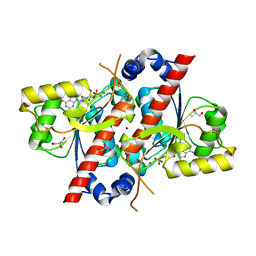 | | Crystal structure of E. coli tRNA N6-threonylcarbamoyladenosine dehydratase, TcdA, in complex with AMP at 1.801 Angstroem resolution | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Lopez-Estepa, M, Arda, A, Savko, M, Round, A, Shepard, W, Bruix, M, Coll, M, Fernandez, F.J, Jimenez-Barbero, J, Vega, M.C. | | Deposit date: | 2014-11-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The Crystal Structure and Small-Angle X-Ray Analysis of Csdl/Tcda Reveal a New tRNA Binding Motif in the Moeb/E1 Superfamily.
Plos One, 10, 2015
|
|
4CX8
 
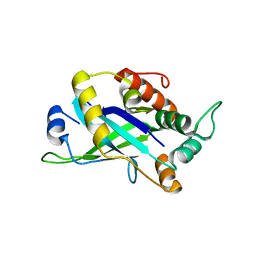 | | Monomeric pseudorabies virus protease pUL26N at 2.5 A resolution | | Descriptor: | PSEUDORABIES VIRUS PROTEASE | | Authors: | Zuehlsdorf, M, Werten, S, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-04 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin Pul26N, a Herpesvirus Serine Protease.
Plos Pathog., 11, 2015
|
|
