4D1D
 
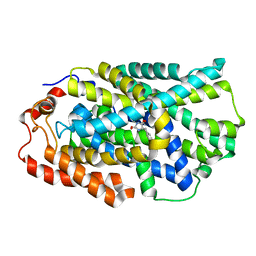 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER with the inhibitor 5-(2-naphthylmethyl)-L-hydantoin. | | Descriptor: | 5-(2-NAPHTHYLMETHYL)-D-HYDANTOIN, 5-(2-NAPHTHYLMETHYL)-L-HYDANTOIN, HYDANTOIN TRANSPORT PROTEIN, ... | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
2Q2T
 
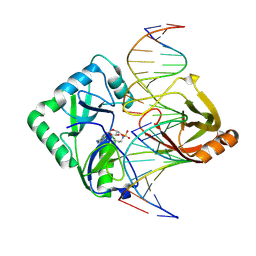 | | Structure of Chlorella virus DNA ligase-adenylate bound to a 5' phosphorylated nick | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', 5'-D(P*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', ... | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q2U
 
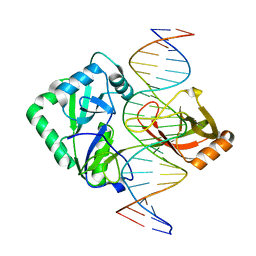 | | Structure of Chlorella virus DNA ligase-product DNA complex | | Descriptor: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', Chlorella virus DNA ligase | | Authors: | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | Deposit date: | 2007-05-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6N0V
 
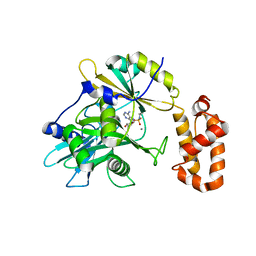 | | tRNA ligase | | Descriptor: | MANGANESE (II) ION, tRNA ligase | | Authors: | Banerjee, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-01-09 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure and two-metal mechanism of fungal tRNA ligase.
Nucleic Acids Res., 47, 2019
|
|
5DKA
 
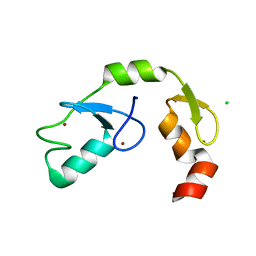 | | A C2HC zinc finger is essential for the activity of the RING ubiquitin ligase RNF125 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF125, MAGNESIUM ION, ... | | Authors: | Boer, D.R, Coll, M, Bijlmakers, M.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A C2HC zinc finger is essential for the RING-E2 interaction of the ubiquitin ligase RNF125.
Sci Rep, 6, 2016
|
|
5HPT
 
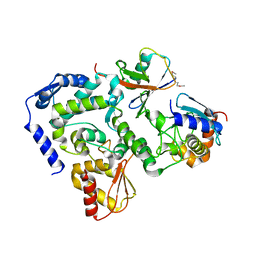 | |
1Z5X
 
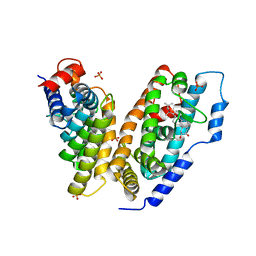 | | hemipteran ecdysone receptor ligand-binding domain complexed with ponasterone A | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor ligand binding domain, PHOSPHATE ION, ... | | Authors: | Carmichael, J.A, Lawrence, M.C, Graham, L.D, Pilling, P.A, Epa, V.C, Noyce, L, Lovrecz, G, Winkler, D.A, Pawlak-Skrzecz, A. | | Deposit date: | 2005-03-21 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | The X-ray structure of a hemipteran ecdysone receptor ligand-binding domain: comparison with a lepidopteran ecdysone receptor ligand-binding domain and implications for insecticide design.
J.Biol.Chem., 280, 2005
|
|
1NWN
 
 | |
7BVB
 
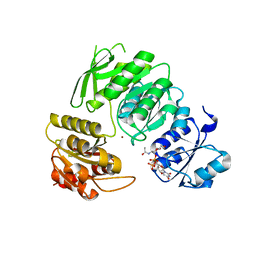 | |
7BVA
 
 | |
1UW0
 
 | |
2HIX
 
 | |
6FYH
 
 | | Disulfide between ubiquitin G76C and the E3 HECT ligase Huwe1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Jaeckl, M, Hartmann, M.D, Wiesner, S. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.906 Å) | | Cite: | beta-Sheet Augmentation Is a Conserved Mechanism of Priming HECT E3 Ligases for Ubiquitin Ligation.
J. Mol. Biol., 430, 2018
|
|
4GLX
 
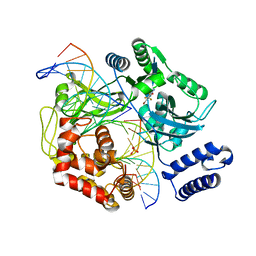 | | DNA ligase A in complex with inhibitor | | Descriptor: | 2-amino-6-bromo-7-(trifluoromethyl)-1,8-naphthyridine-3-carboxamide, DNA (26-MER), DNA (5'-D(*AP*CP*AP*AP*TP*TP*GP*CP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Prade, L, Lange, R, Tidten-Luksch, N, Chambovey, A. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4GLW
 
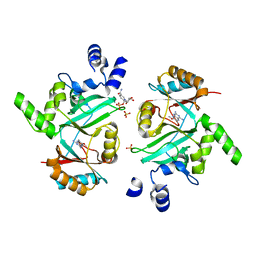 | | DNA ligase A in complex with inhibitor | | Descriptor: | 7-methoxy-6-methylpteridine-2,4-diamine, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Prade, L, Lange, R, Tidten-Luksch, N, Chambovey, A. | | Deposit date: | 2012-08-15 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2HIV
 
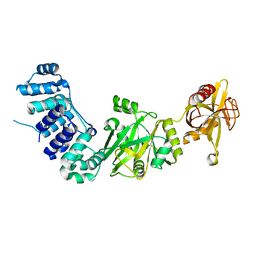 | | ATP-dependent DNA ligase from S. solfataricus | | Descriptor: | Thermostable DNA ligase | | Authors: | Pascal, J.M, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2QVY
 
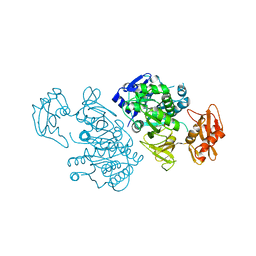 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3,4-Dichlorobenzoate | | Descriptor: | 3,4-dichlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
2QVX
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, I303G mutation, bound to 3-Chlorobenzoate | | Descriptor: | 3-chlorobenzoate, 4-Chlorobenzoate CoA Ligase | | Authors: | Wu, R, Reger, A.S, Cao, J, Gulick, A.M, Dunaway-Mariano, D. | | Deposit date: | 2007-08-09 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Rational redesign of the 4-chlorobenzoate binding site of 4-chlorobenzoate: coenzyme a ligase for expanded substrate range.
Biochemistry, 46, 2007
|
|
6J1Y
 
 | | Semi-open conformation E3 ligase | | Descriptor: | GLYCEROL, NEDD4-like E3 ubiquitin-protein ligase WWP1 | | Authors: | Liu, Z.H. | | Deposit date: | 2018-12-30 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.
Nat Commun, 10, 2019
|
|
4IO5
 
 | |
6JOM
 
 | | Crystal structure of lipoate protein ligase from Mycoplasma hyopneumoniae | | Descriptor: | 5'-O-[(R)-({5-[(3R)-1,2-DITHIOLAN-3-YL]PENTANOYL}OXY)(HYDROXY)PHOSPHORYL]ADENOSINE, Lipoate--protein ligase | | Authors: | Zhang, H, Chen, H, Ma, G. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Functional Identification and Structural Analysis of a New Lipoate Protein Ligase inMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 10, 2020
|
|
3CW9
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase in the Thioester-forming Conformation, bound to 4-chlorophenacyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-Chlorophenacyl-coenzyme A, 4-chlorobenzoyl CoA ligase, ... | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
4IO3
 
 | |
1P3D
 
 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
3CW8
 
 | | 4-Chlorobenzoyl-CoA Ligase/Synthetase, bound to 4CBA-Adenylate | | Descriptor: | 1,2-ETHANEDIOL, 4-chlorobenzoyl CoA ligase, 5'-O-[(S)-{[(4-chlorophenyl)carbonyl]oxy}(hydroxy)phosphoryl]adenosine | | Authors: | Reger, A.S, Cao, J, Wu, R, Dunaway-Mariano, D, Gulick, A.M. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterization of a 140 degrees domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase.
Biochemistry, 47, 2008
|
|
