5IAJ
 
 | | Caspase 3 V266L | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4C80
 
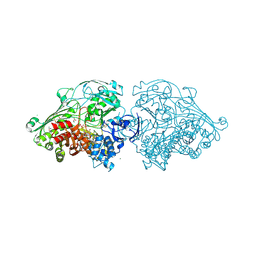 | | Aldehyde Oxidoreductase from Desulfovibrio gigas (MOP), soaked with hydrogen peroxide | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), ALDEHYDE OXIDOREDUCTASE, BICARBONATE ION, ... | | Authors: | Correia, H.D, Romao, M.J, Santos-Silva, T. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic and Structural Studies of Aldehyde Oxidoreductase from Desulfovibrio Gigas Reveal a Dithiolene-Based Chemistry for Enzyme Activation and Inhibition by H2O2.
Plos One, 8, 2013
|
|
5INB
 
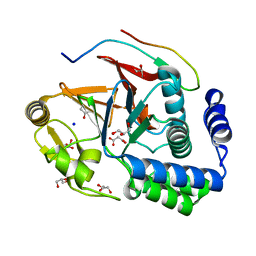 | | RepoMan-PP1g (protein phosphatase 1, gamma isoform) holoenzyme complex | | Descriptor: | Cell division cycle-associated protein 2, GLYCEROL, MALONATE ION, ... | | Authors: | Kumar, G.S, Peti, W, Page, R. | | Deposit date: | 2016-03-07 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Elife, 5, 2016
|
|
1P54
 
 | |
5IR5
 
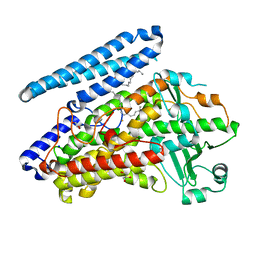 | | Crystal structure of wild-type bacterial lipoxygenase from Pseudomonas aeruginosa PA-LOX with space group P21212 at 1.9 A resolution | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, FE (II) ION, ... | | Authors: | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa.
Biochim.Biophys.Acta, 1861, 2016
|
|
1OUA
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE I56T MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Ogasahara, K, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure, stability, and folding process of amyloidogenic mutant human lysozyme.
J.Biochem.(Tokyo), 120, 1996
|
|
1OUI
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V93A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
7A0X
 
 | | Structure of dimeric sodium proton antiporter NhaA, at pH 6.0, crystallized with chimeric Fab antibodies | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Fippel, A, Lentes, C.J, Mir, S.H, Wirth, C, Hunte, C. | | Deposit date: | 2020-08-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Molecular determinants for substrate uptake in electrogenic sodium/proton antiporters
To Be Published
|
|
7A0W
 
 | | Structure of dimeric sodium proton antiporter NhaA, at pH 8.5, crystallized with chimeric Fab antibodies | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CARDIOLIPIN, CHLORIDE ION, ... | | Authors: | Fippel, A, Gross, N.M, Mir, S.H, Wirth, C, Hunte, C. | | Deposit date: | 2020-08-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Molecular determinants for substrate uptake in electrogenic sodium/proton antiporters
To Be Published
|
|
7A0Y
 
 | | Structure of dimeric sodium proton antiporter NhaA K300R variant, at pH 8.2, crystallized with chimeric Fab antibodies | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Fippel, A, Mir, S.H, Lentes, C.J, Wirth, C, Hunte, C. | | Deposit date: | 2020-08-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Molecular determinants for substrate uptake in electrogenic sodium/proton antiporters
To Be Published
|
|
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5I9B
 
 | | Caspase 3 V266A | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4AZY
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND 10) | | Descriptor: | (1S)-4-fluoro-1-(4-fluoro-3-pyrimidin-5-ylphenyl)-1-[2-(trifluoromethyl)pyridin-4-yl]-1H-isoindol-3-amine, ACETATE ION, BETA-SECRETASE 1, ... | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
5IBC
 
 | | Caspase 3 V266I | | Descriptor: | ACE-ASP-GLU-VAL-ASK, Caspase-3, SODIUM ION | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2TYS
 
 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | Descriptor: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | Authors: | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
5IAR
 
 | | Caspase 3 V266W | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACE-ASP-GLU-VAL-ASK, Caspase-3, ... | | Authors: | Maciag, J.J, Mackenzie, S.H, Tucker, M.B, Schipper, J.L, Swartz, P.D, Clark, A.C. | | Deposit date: | 2016-02-21 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Tunable allosteric library of caspase-3 identifies coupling between conserved water molecules and conformational selection.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IBW
 
 | | Complex of MlcC bound to the tandem IQ motif of MyoC | | Descriptor: | Calcium-binding EF-hand domain-containing protein, Myosin IC heavy chain, SODIUM ION | | Authors: | Langelaan, D.N, Smith, S.P. | | Deposit date: | 2016-02-22 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Single-lobe Myosin Light Chain C in Complex with the Light Chain-binding Domains of Myosin-1C Provides Insights into Divergent IQ Motif Recognition.
J.Biol.Chem., 291, 2016
|
|
5ICF
 
 | | Crystal structure of (S)-norcoclaurine 6-O-methyltransferase with S-adenosyl-L-homocysteine and sanguinarine | | Descriptor: | (S)-norcoclaurine 6-O-methyltransferase, 1,2-ETHANEDIOL, 13-methyl[1,3]benzodioxolo[5,6-c][1,3]dioxolo[4,5-i]phenanthridin-13-ium, ... | | Authors: | Robin, A.Y, Graindorge, M, Giustini, C, Dumas, R, Matringe, M. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids.
Plant J., 87, 2016
|
|
3AG0
 
 | | Crystal structure of carbonmonoxy human cytoglobin | | Descriptor: | CARBON MONOXIDE, Cytoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sawai, H, Shiro, Y, Sugimoto, H. | | Deposit date: | 2010-03-17 | | Release date: | 2011-02-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the carbon monoxide complex of human cytoglobin
Proteins, 79, 2011
|
|
5IHH
 
 | | Crystal structure of human transthyretin in complex with luteolin-MeO at 1.35 A resolution | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5-hydroxy-7-methoxy-4H-1-chromen-4-one, SODIUM ION, Transthyretin | | Authors: | Begum, A, Nilsson, L, Olofsson, A, Sauer-Eriksson, A.E. | | Deposit date: | 2016-02-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity.
Plos One, 11, 2016
|
|
1WPC
 
 | | Crystal structure of maltohexaose-producing amylase complexed with pseudo-maltononaose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2004-09-01 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and crystallographic analyses of maltohexaose-producing amylase from alkalophilic Bacillus sp. 707
Biochemistry, 43, 2004
|
|
4DKA
 
 | | Structure of Editosome protein | | Descriptor: | RNA-editing complex protein MP81, SODIUM ION, single domain antibody VHH | | Authors: | Park, Y.-J, Hol, W. | | Deposit date: | 2012-02-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the C-terminal domain of the largest editosome interaction protein and its role in promoting RNA binding by RNA-editing ligase L2.
Nucleic Acids Res., 40, 2012
|
|
5IHG
 
 | | The X-ray structure of the adduct formed in the reaction between hen egg white lysozyme a compound I, a platin(II) compound containing a O, S bidentate ligand | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Ferraro, G, Merlino, A. | | Deposit date: | 2016-02-29 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Platinum(ii) O,S complexes as potential metallodrugs against Cisplatin resistance.
Dalton Trans, 45, 2016
|
|
3CLO
 
 | |
4DC4
 
 | | Lysozyme Trimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
