4C1X
 
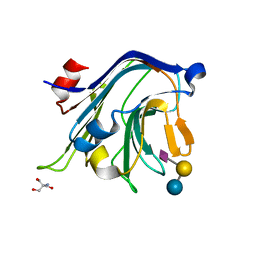 | | Carbohydrate binding domain from Streptococcus pneumoniae NanA sialidase complexed with 6'-sialyllactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, SIALIDASE A | | Authors: | Yang, L, Connaris, H, Potter, J.A, Taylor, G.L. | | Deposit date: | 2013-08-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Characterization of the Carbohydrate-Binding Module of Nana Sialidase, a Pneumococcal Virulence Factor.
Bmc Struct.Biol., 15, 2015
|
|
4RUX
 
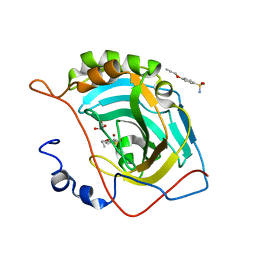 | |
6MZZ
 
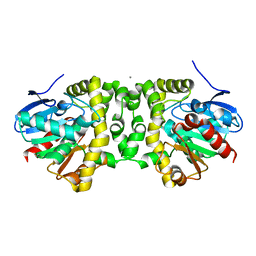 | | Fluoroacetate dehalogenase, room temperature structure, using first 1 degree of total 3 degree oscillation | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
4TLJ
 
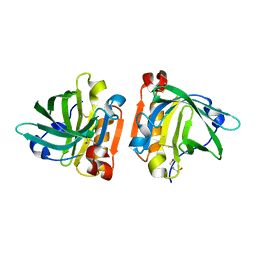 | | Ultra-high resolution crystal structure of caprine Beta-lactoglobulin | | Descriptor: | 1,4-BUTANEDIOL, Beta-lactoglobulin | | Authors: | Crowther, J.M, Jameson, G.B, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ultra-high resolution crystal structure of recombinant caprine beta-lactoglobulin.
Febs Lett., 588, 2014
|
|
7GEL
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAG-UCB-cedd89ab-1 (Mpro-x11475) | | Descriptor: | (1M,3P)-1-(3-chlorophenyl)-3-(4-methylpyridin-3-yl)-1,3-dihydro-2H-imidazol-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5HOS
 
 | |
3OWC
 
 | | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, Probable acetyltransferase | | Authors: | Majorek, K.A, Chruszcz, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of GNAT superfamily protein PA2578 from Pseudomonas aeruginosa
To be Published
|
|
5DMU
 
 | | Structure of the NHEJ polymerase from Methanocella paludicola | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NHEJ Polymerase, ... | | Authors: | Brissett, N.C, Bartlett, E.J, Doherty, A.J. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Molecular basis for DNA strand displacement by NHEJ repair polymerases.
Nucleic Acids Res., 44, 2016
|
|
4TMT
 
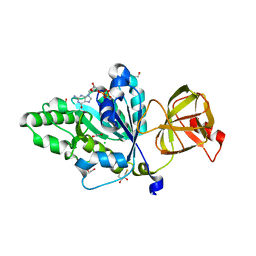 | | Translation initiation factor eIF5B (517-858) mutant D533A from C. thermophilum, bound to GTPgammaS | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Kuhle, B, Ficner, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A monovalent cation acts as structural and catalytic cofactor in translational GTPases.
Embo J., 33, 2014
|
|
6MWE
 
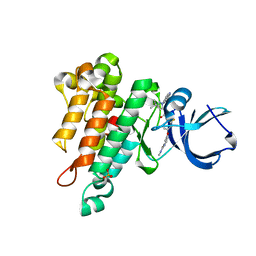 | | CRYSTAL STRUCTURE OF TIE2 IN COMPLEX WITH DECIPERA COMPOUND DP1919 | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, Angiopoietin-1 receptor, SULFATE ION | | Authors: | Chun, L, Abendroth, J, Edwards, T.E. | | Deposit date: | 2018-10-29 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Selective Tie2 Inhibitor Rebastinib Blocks Recruitment and Function of Tie2
Mol. Cancer Ther., 16, 2017
|
|
4CD7
 
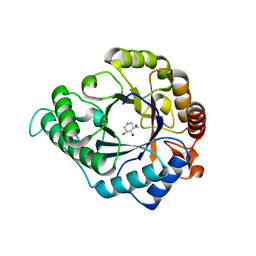 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG and beta-1,4-mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
5U3Q
 
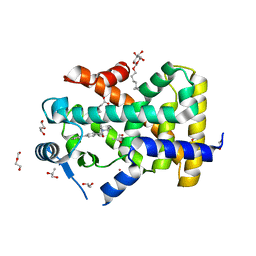 | | Human PPARdelta ligand-binding domain in complexed with specific agonist 1 | | Descriptor: | 6-(2-{[([1,1'-biphenyl]-4-carbonyl)(propan-2-yl)amino]methyl}phenoxy)hexanoic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, C.-C, Baiga, T.J, Downes, M, La Clair, J.J, Atkins, A.R, Richard, S.B, Stockley-Noel, T.A, Bowman, M.E, Evans, R.M, Noel, J.P. | | Deposit date: | 2016-12-03 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7GD8
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with JAG-UCB-119787ef-1 (Mpro-x10898) | | Descriptor: | (4R)-6-chloro-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4XGN
 
 | |
6FFX
 
 | | Crystal structure of R. ruber ADH-A, mutant F43H | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2018-01-09 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed Evolution of Alcohol Dehydrogenase for Improved Stereoselective Redox Transformations of 1-Phenylethane-1,2-diol and Its Corresponding Acyloin.
Biochemistry, 57, 2018
|
|
4XFJ
 
 | |
3KTL
 
 | | Crystal Structure of an I71A human GSTA1-1 mutant in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Achilonu, I.A, Gildenhuys, S, Fernandes, M.A, Fanucchi, S, Dirr, H.W. | | Deposit date: | 2009-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The role of a topologically conserved isoleucine in glutathione transferase structure, stability and function.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4C2Z
 
 | | Human N-myristoyltransferase (NMT1) with Myristoyl-CoA and inhibitor bound | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Thinon, E, Serwa, R.A, Brannigan, J.A, Brassat, U, Wright, M.H, Heal, W.P, Wilkinson, A.J, Mann, D.J, Tate, E.W. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chemical Proteomics Defines the Mammalian N- Myristoylated Proteome in Live Cells Global Profiling of Co- and Post-Translationally N-Myristoylated Proteomes in Human Cells.
Nat.Commun., 5, 2014
|
|
5DSP
 
 | |
6FH7
 
 | | Crystal Structure of BAZ2B bromodomain in complex with 1-methylpyridinone compound 2 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, ~{N}-[3-[(4-fluorophenyl)carbonylamino]propyl]-1-methyl-6-oxidanylidene-pyridine-3-carboxamide | | Authors: | Dalle Vedove, A, Spiliotopoulos, D, Lolli, G, Caflisch, A. | | Deposit date: | 2018-01-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Small-Molecule Binding to the BAZ2A and BAZ2B Bromodomains.
ChemMedChem, 13, 2018
|
|
6F8U
 
 | | Crystal structure of the PDE4D catalytic domain in complex with GEBR-20b | | Descriptor: | 2-[(~{E})-[4-[bis(fluoranyl)methoxy]-3-cyclopentyloxy-phenyl]methylideneamino]oxy-1-[(2~{R},6~{R})-2,6-dimethylmorpholin-4-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Prosdocimi, T, Donini, S, Parisini, E. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Bases of PDE4D Inhibition by Memory-Enhancing GEBR Library Compounds.
Biochemistry, 57, 2018
|
|
3KWF
 
 | | human DPP-IV with carmegliptin (S)-1-((2S,3S,11bS)-2-Amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl)-4-fluoromethyl-pyrrolidin-2-one | | Descriptor: | (4S)-1-[(2S,3S,11bS)-2-amino-9,10-dimethoxy-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-3-yl]-4-(fluoromethyl)pyrrolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4 | | Authors: | Hennig, M, Stihle, M, Thoma, R. | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of carmegliptin: A potent and long-acting dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6CJI
 
 | | Candida albicans Hsp90 nucleotide binding domain | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 90 homolog | | Authors: | Hutchinson, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Pizarro, J.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
7GFV
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with MAT-POS-2db6411e-2 (Mpro-x11852) | | Descriptor: | 1-{4-[(3-chloro-5-hydroxyphenyl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
7GG5
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-87c86d55-1 (Mpro-x12143) | | Descriptor: | 2-(1H-benzotriazol-1-yl)-N-[4-(dimethylamino)phenyl]-N-[(1,3-thiazol-4-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.509 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
