1NWP
 
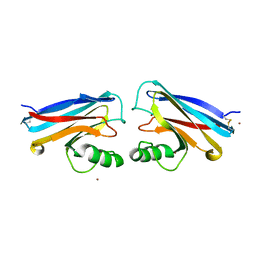 | |
1HUS
 
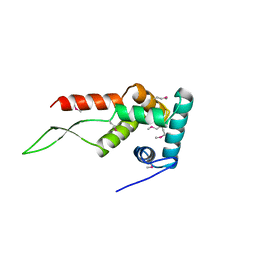 | | RIBOSOMAL PROTEIN S7 | | Descriptor: | RIBOSOMAL PROTEIN S7 | | Authors: | Hosaka, H, Nakagawa, A, Tanaka, I. | | Deposit date: | 1997-08-08 | | Release date: | 1998-01-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ribosomal protein S7: a new RNA-binding motif with structural similarities to a DNA architectural factor.
Structure, 5, 1997
|
|
1FSV
 
 | |
1F3Z
 
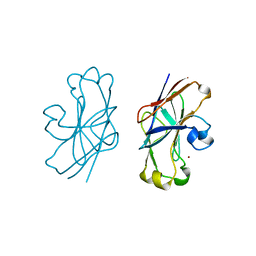 | | IIAGLC-ZN COMPLEX | | Descriptor: | GLUCOSE-SPECIFIC PHOSPHOCARRIER, ZINC ION | | Authors: | Feese, M, Comolli, L, Meadow, N, Roseman, S, Remington, S.J. | | Deposit date: | 1997-10-09 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural studies of the Escherichia coli signal transducing protein IIAGlc: implications for target recognition.
Biochemistry, 36, 1997
|
|
1AXI
 
 | | STRUCTURAL PLASTICITY AT THE HGH:HGHBP INTERFACE | | Descriptor: | GROWTH HORMONE, GROWTH HORMONE RECEPTOR, SULFATE ION | | Authors: | Atwell, S, Ultsch, M, De Vos, A.M, Wells, J.A. | | Deposit date: | 1997-10-15 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural plasticity in a remodeled protein-protein interface.
Science, 278, 1997
|
|
3JDW
 
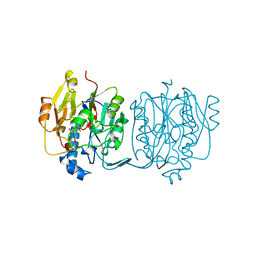 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
2HEB
 
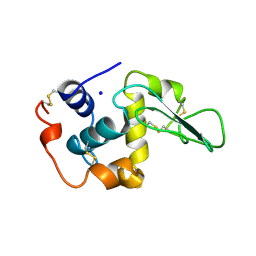 | | CONTRIBUTION OF WATER MOLECULES IN THE INTERIOR OF A PROTEIN TO THE CONFORMATIONAL STABILITY | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Funahashi, J, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of water molecules in the interior of a protein to the conformational stability.
J.Mol.Biol., 274, 1997
|
|
1AL0
 
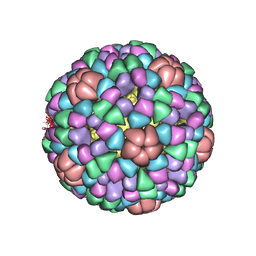 | | PROCAPSID OF BACTERIOPHAGE PHIX174 | | Descriptor: | CAPSID PROTEIN GPF, SCAFFOLDING PROTEIN GPB, SCAFFOLDING PROTEIN GPD, ... | | Authors: | Rossmann, M.G, Dokland, T. | | Deposit date: | 1997-06-06 | | Release date: | 1998-01-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of a viral procapsid with molecular scaffolding.
Nature, 389, 1997
|
|
1AA7
 
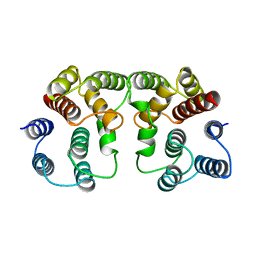 | |
1PSV
 
 | |
1ZNJ
 
 | | INSULIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, INSULIN, PHENOL, ... | | Authors: | Turkenburg, M.G.W, Whittingham, J.L, Turkenburg, J.P, Dodson, G.G, Derewenda, U, Smith, G.D, Dodson, E.J, Derewenda, Z.S, Xiao, B. | | Deposit date: | 1997-09-23 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure Determination and Refinement of Two Crystal Forms of Native Insulins
To be Published
|
|
1BJE
 
 | |
1XLM
 
 | | D254E, D256E MUTANT OF D-XYLOSE ISOMERASE COMPLEXED WITH AL3 AND XYLITOL | | Descriptor: | ALUMINUM ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Gerczei, T, Bocskei, Z.S, Szabo, E, Naray-Szabo, G, Asboth, B. | | Deposit date: | 1997-07-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure determination and refinement of the Al3+ complex of the D254,256E mutant of Arthrobacter D-xylose isomerase at 2.40 A resolution. Further evidence for inhibitor-induced metal ion movement.
Int.J.Biol.Macromol., 25, 1999
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
4GAT
 
 | | SOLUTION NMR STRUCTURE OF THE WILD TYPE DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13BP DNA CONTAINING A CGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*GP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors.
J.Mol.Biol., 277, 1998
|
|
2GAT
 
 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF CHICKEN GATA-1 BOUND TO DNA, NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*TP*CP*TP*GP*CP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*GP*CP*AP*GP*AP*TP*AP*AP*AP*CP*AP*TP*T)-3'), ERYTHROID TRANSCRIPTION FACTOR GATA-1, ... | | Authors: | Clore, G.M, Tjandra, N, Starich, M, Omichinski, J.G, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Use of dipolar 1H-15N and 1H-13C couplings in the structure determination of magnetically oriented macromolecules in solution.
Nat.Struct.Biol., 4, 1997
|
|
1AQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1997-08-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
1AP5
 
 | |
1ARK
 
 | |
1AQB
 
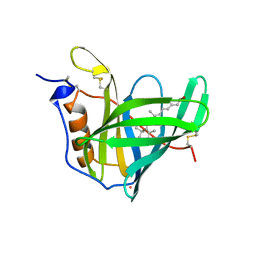 | | RETINOL-BINDING PROTEIN (RBP) FROM PIG PLASMA | | Descriptor: | CADMIUM ION, RETINOL, RETINOL-BINDING PROTEIN | | Authors: | Zanotti, G, Panzalorto, M, Marcato, A, Malpeli, G, Folli, C, Berni, R. | | Deposit date: | 1997-07-28 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of pig plasma retinol-binding protein at 1.65 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1AU7
 
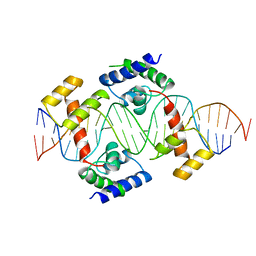 | | PIT-1 MUTANT/DNA COMPLEX | | Descriptor: | CONSENSUS DNA 25-MER, DNA (5'-D(*CP*TP*TP*CP*CP*TP*CP*AP*TP*GP*TP*AP*TP*AP*TP*AP*C P*AP*TP*GP*AP*GP* GP*A)-3'), PROTEIN PIT-1 | | Authors: | Jacobson, E.M, Li, P, Leon-Del-Rio, A, Rosenfeld, M.G, Aggarwal, A.K. | | Deposit date: | 1997-09-12 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Pit-1 POU domain bound to DNA as a dimer: unexpected arrangement and flexibility.
Genes Dev., 11, 1997
|
|
1AYJ
 
 | |
1G3P
 
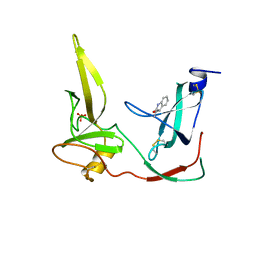 | | CRYSTAL STRUCTURE OF THE N-TERMINAL DOMAINS OF BACTERIOPHAGE MINOR COAT PROTEIN G3P | | Descriptor: | MINOR COAT PROTEIN, SULFATE ION | | Authors: | Lubkowski, J, Hennecke, F, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1997-12-22 | | Release date: | 1998-01-28 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The structural basis of phage display elucidated by the crystal structure of the N-terminal domains of g3p.
Nat.Struct.Biol., 5, 1998
|
|
1CKO
 
 | |
6GAT
 
 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
