7YD0
 
 | |
5TF0
 
 | | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis | | Descriptor: | 1,2-ETHANEDIOL, Glycosyl hydrolase family 3 N-terminal domain protein, MAGNESIUM ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-23 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2021 Å) | | Cite: | Crystal Structure of Glycosil Hydrolase Family 3 N-Terminal Domain Protein from Bacteroides intestinalis
To Be Published, 2016
|
|
3WH5
 
 | | Crystal structure of GH1 beta-glucosidase Td2F2 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Jo, T, Fushinobu, S, Uchiyama, T, Yaoi, K. | | Deposit date: | 2013-08-21 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure and identification of a key amino acid for glucose tolerance, substrate specificity, and transglycosylation activity of metagenomic beta-glucosidase Td2F2.
Febs J., 283, 2016
|
|
7FZM
 
 | | Crystal Structure of human FABP4 in complex with N-(2,4-dichlorophenyl)-2-(2,6-dihydroxypyrimidin-4-yl)acetamide, i.e. SMILES n1c(cc(nc1O)CC(=O)Nc1c(cc(cc1)Cl)Cl)O with IC50=15 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, N-(2,4-dichlorophenyl)-2-(2,6-dioxo-1,2,5,6-tetrahydropyrimidin-4-yl)acetamide, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3HCE
 
 | | Crystal Structure of E185D hPNMT in Complex With Octopamine and AdoHcy | | Descriptor: | 4-(2R-AMINO-1-HYDROXYETHYL)PHENOL, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity.
Biochem.J., 422, 2009
|
|
4OW5
 
 | | Structural basis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles | | Descriptor: | 1,2-ETHANEDIOL, Fusolin | | Authors: | Hijnen, M, Boudes, M, Aizel, K, Coulibaly, F. | | Deposit date: | 2014-01-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
5E3B
 
 | | Structure of macrodomain protein from Streptomyces coelicolor | | Descriptor: | 1,2-ETHANEDIOL, Macrodomain protein, SODIUM ION | | Authors: | Lalic, J, Posavec Marjanovic, M, Perina, D, Sabljic, I, Zaja, R, Plese, B, Imesek, M, Bucca, G, Ahel, M, Cetkovic, H, Luic, M, Mikoc, A, Ahel, I. | | Deposit date: | 2015-10-02 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Disruption of Macrodomain Protein SCO6735 Increases Antibiotic Production in Streptomyces coelicolor.
J.Biol.Chem., 291, 2016
|
|
5UV4
 
 | | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Putative leucine-rich repeat protein kinase family protein | | Authors: | Counago, R.M, Aquino, B, Massirer, K.B, Gileadi, O, Arruda, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-02-17 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Maize SIRK1 (sucrose-induced receptor kinase 1) kinase domain bound to AMP-PNP
To Be Published
|
|
5U9K
 
 | |
7JWI
 
 | |
4GKZ
 
 | | HA1.7, a MHC class II restricted TCR specific for haemagglutinin | | Descriptor: | 1,2-ETHANEDIOL, Alpha chain of Class II TCR, Beta Chain of Class II TCR, ... | | Authors: | Holland, C.J, Rizkallah, P.J, Cole, D.K, Sewell, A.K, Godkin, A.J. | | Deposit date: | 2012-08-13 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Minimal conformational plasticity enables TCR cross-reactivity to different MHC class II heterodimers.
Sci Rep, 2, 2012
|
|
2YT8
 
 | | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma) | | Descriptor: | Amyloid beta A4 precursor protein-binding family A member 3 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Tarada, T, Seiki, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-05 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PDZ domain of Amyloid beta A4 precursor protein-binding family A member 3 (Neuron- specific X11L2 protein) (Neuronal Munc18-1-interacting protein 3) (Mint-3) (Adapter protein X11gamma)
To be Published
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5U87
 
 | | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin | | Descriptor: | Preproalbumin PawS1 | | Authors: | Franke, B, James, A.M, Mobli, M, Colgrave, M.L, Mylne, J.S, Rosengren, K.J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the precursor protein PawS1 comprising SFTI-1 and a seed storage albumin
to be published
|
|
3W8R
 
 | | Mutant structure of Thermus thermophilus HB8 uridine-cytidine kinase | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Uridine kinase | | Authors: | Tomoike, F, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2013-03-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Indispensable residue for uridine binding in the uridine-cytidine kinase family.
Biochem Biophys Rep, 11, 2017
|
|
3QW1
 
 | |
1ND3
 
 | | The structure of HRV16, when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, ZINC ION, coat protein VP1, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-06 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
4D0C
 
 | | COMPLEX OF A B21 CHICKEN MHC CLASS I MOLECULE AND A 10MER CHICKEN PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 10MER PEPTIDE, BETA-2-MICROGLOBULIN, ... | | Authors: | Chappell, P.E, Roversi, P, Harrison, M.C, Mears, L.E, Kaufman, J.F, Lea, S.M. | | Deposit date: | 2014-04-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Expression levels of MHC class I molecules are inversely correlated with promiscuity of peptide binding.
Elife, 4, 2015
|
|
3GPI
 
 | | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus | | Descriptor: | 1,2-ETHANEDIOL, NAD-dependent epimerase/dehydratase | | Authors: | Ramagopal, U.A, Morano, C, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-23 | | Release date: | 2009-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structure of putative NAD-dependent epimerase/dehydratase from methylobacillus flagellatus
To be published
|
|
1NCR
 
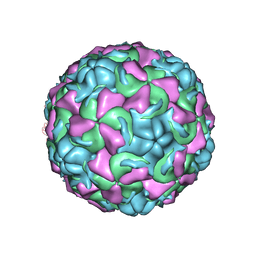 | | The structure of Rhinovirus 16 when complexed with pleconaril, an antiviral compound | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, MYRISTIC ACID, ZINC ION, ... | | Authors: | Zhang, Y, Simpson, A.A, Bator, C.M, Chakravarty, S, Pevear, D.C, Skochko, G.A, Tull, T.M, Diana, G, Rossmann, M.G. | | Deposit date: | 2002-12-05 | | Release date: | 2003-12-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds
J.Virol., 78, 2004
|
|
4U0V
 
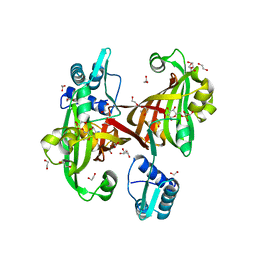 | |
4KIL
 
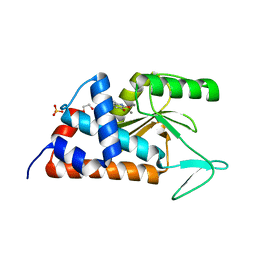 | | 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 3-Hydroxyquinolin-2(1H)-ones As Inhibitors of Influenza A Endonuclease.
ACS Med Chem Lett, 4, 2013
|
|
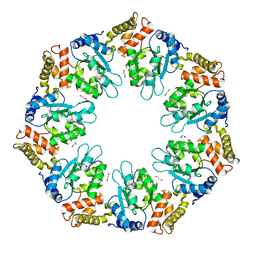
6LUA
 
 | |
1K6F
 
 | |
