7ZM8
 
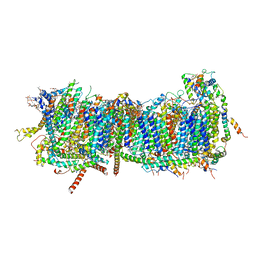 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (inhibited by DDM) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7ZME
 
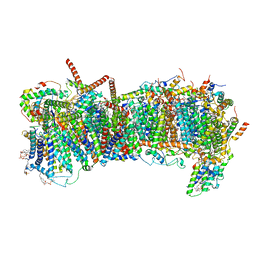 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
8JTR
 
 | | Cryo-EM structure of GeoCas9-sgRNA binary complex | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA (139-bp) | | Authors: | Shen, P.P, Liu, B.B, Li, X, Zhang, L.L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-06-22 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of GeoCas9-sgRNA binary complex
To Be Published
|
|
6J8H
 
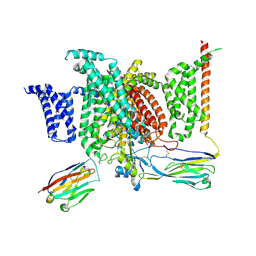 | | Structure of human voltage-gated sodium channel Nav1.7 in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (Y1755 down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sodium channel protein type 9 subunit alpha, ... | | Authors: | Shen, H, Liu, D, Lei, J, Yan, N. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins.
Science, 363, 2019
|
|
5A09
 
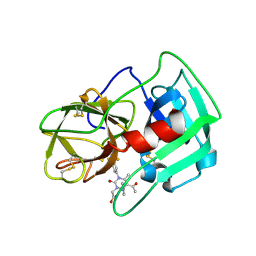 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-[(4R)-4-(4-cyanophenyl)-5-ethanoyl-6-methyl-2-oxidanylidene-1-[3-(trifluoromethyl)phenyl]-4H-pyrimidin-3-yl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEUTROPHIL ELASTASE, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
1D8E
 
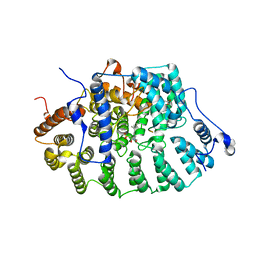 | | Zinc-depleted FTase complexed with K-RAS4B peptide substrate and FPP analog. | | Descriptor: | ACETATE ION, FARNESYLTRANSFERASE (ALPHA SUBUNIT), FARNESYLTRANSFERASE (BETA SUBUNIT), ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 1999-10-22 | | Release date: | 2000-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The basis for K-Ras4B binding specificity to protein farnesyltransferase revealed by 2 A resolution ternary complex structures.
Structure Fold.Des., 8, 2000
|
|
5A0A
 
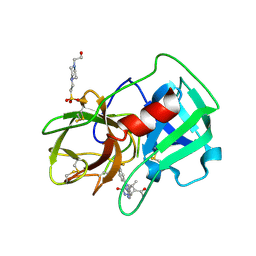 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-[(6R)-5-ethanoyl-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidin-6-yl]pyridine-2-carbonitrile, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
4GY1
 
 | |
8JVZ
 
 | |
8JVN
 
 | |
8JVT
 
 | |
8JVY
 
 | |
8JVS
 
 | |
8JVR
 
 | |
8JVU
 
 | |
8JVW
 
 | |
5A0C
 
 | | Crystal Structure of human neutrophil elastase in complex with a dihydropyrimidone inhibitor | | Descriptor: | (6S)-6-(4-cyano-2-methylsulfonyl-phenyl)-4-methyl-2-oxidanylidene-3-[3-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-5-carbonitrile, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | vonNussbaum, F, Li, V.M.-J, Allerheiligen, S, Anlauf, S, Baerfacker, L, Bechem, M, Delbeck, M, Fitzgerald, M.F, Gerisch, M, Gielen-Haertwig, H, Haning, H, Karthaus, D, Lang, D, Lustig, K, Meibom, D, Mittendorf, J, Rosentreter, U, Schaefer, M, Schaefer, S, Schamberger, J, Telan, L.A, Tersteegen, A. | | Deposit date: | 2015-04-17 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Freezing the Bioactive Conformation to Boost Potency: The Identification of BAY 85-8501, a Selective and Potent Inhibitor of Human Neutrophil Elastase for Pulmonary Diseases.
ChemMedChem, 10, 2015
|
|
8I8N
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Dephosphocoenzyme-A and Phosphonoacetic acid at 2.22 A resolution.
To Be Published
|
|
5FNZ
 
 | |
8I8J
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.07 A resolution.
To Be Published
|
|
8I8K
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.13 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.13 A resolution.
To Be Published
|
|
8I8L
 
 | | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.23 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, GLYCEROL, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structure of the ternary complex of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter sp. with Coenzyme-A and Phosphonoacetic acid at 2.23 A resolution.
To Be Published
|
|
8ING
 
 | | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ahmad, M.I, Viswanathan, V, Kumar, M, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-03-09 | | Release date: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the ternary complex of lactoperoxidase with substrate nitric oxide (NO) and product nitrite ion (NO2) at 1.98 A resolution
To be published
|
|
8JVP
 
 | |
8JVQ
 
 | |
