8UWV
 
 | |
6UTB
 
 | |
6USW
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
6OP3
 
 | |
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
8SBB
 
 | | Cryo-EM structure of FtAlkB | | Descriptor: | Alkane 1-monooxygenase, DODECANE, FE (III) ION, ... | | Authors: | Zhang, J, Feng, L. | | Deposit date: | 2023-04-03 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structure and mechanism of the alkane-oxidizing enzyme AlkB.
Nat Commun, 14, 2023
|
|
6UTD
 
 | |
1SKK
 
 | | Structure of the antimicrobial hexapeptide cyc-(KKWWKF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide KKWWKF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
1SKI
 
 | | Structure of the antimicrobial hexapeptide cyc-(RRYYRF) bound to DPC micelles | | Descriptor: | cyclic hexapeptide RRYYRF | | Authors: | Appelt, C, Soderhall, J.A, Bienert, M, Dathe, M, Schmieder, P. | | Deposit date: | 2004-03-05 | | Release date: | 2005-03-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogues in solution and bound to detergent micelles.
Chembiochem, 6, 2005
|
|
7QHO
 
 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (as isolated) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex
Nature Communications, 13, 2022
|
|
7QHM
 
 | | Cytochrome bcc-aa3 supercomplex (respiratory supercomplex III2/IV2) from Corynebacterium glutamicum (stigmatellin and azide bound) | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, AZIDE ION, ... | | Authors: | Kao, W.-C, Hunte, C. | | Deposit date: | 2021-12-13 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for safe and efficient energy conversion in a respiratory supercomplex.
Nat Commun, 13, 2022
|
|
6QF1
 
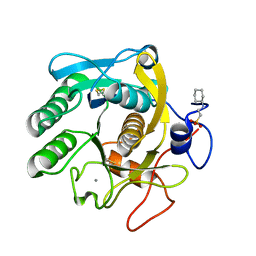 | | X-Ray structure of Proteinase K crystallized on a silicon chip | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CALCIUM ION, Proteinase K | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6D1V
 
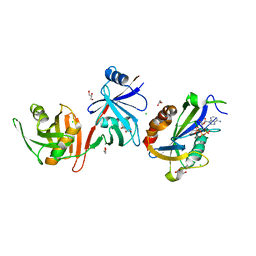 | |
6WEX
 
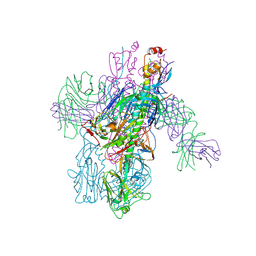 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H6 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
3KPX
 
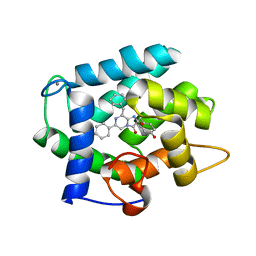 | | Crystal Structure Analysis of photoprotein clytin | | Descriptor: | Apophotoprotein clytin-3, C2-HYDROPEROXY-COELENTERAZINE, CALCIUM ION | | Authors: | Titushin, M.S, Li, Y, Stepanyuk, G.A, Wang, B.-C, Lee, J, Vysotski, E.S, Liu, Z.-J. | | Deposit date: | 2009-11-17 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | NMR derived topology of a GFP-photoprotein energy transfer complex
J.Biol.Chem., 285, 2010
|
|
6WF1
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H10 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, Hemagglutinin HA2 chain, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.19 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6QF2
 
 | | X-Ray structure of Thermolysin crystallized on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CALCIUM ION, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6WF0
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14 Bound to the Influenza A H3 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
6QF3
 
 | | X-Ray structure of Thermolysin soaked with sodium aspartate on a silicon chip | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ASPARTIC ACID, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6WEZ
 
 | | Crystal Structure of Broadly Neutralizing Antibody 3I14-D93N Mutant Bound to the Influenza A H3 Hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hemagglutinin, ... | | Authors: | Harshbarger, W.D, Lockbaum, G.J, Deming, D.T, Attatippaholkun, N, Schiffer, C.A, Marasco, W.A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Unique structural solution from a V H 3-30 antibody targeting the hemagglutinin stem of influenza A viruses.
Nat Commun, 12, 2021
|
|
4B2S
 
 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
6QF4
 
 | | X-Ray structure of human Serine/Threonine Kinase 17B (STK17B) aka DRAK2 in complex with ADP obtained by on-chip soaking | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
6QF5
 
 | | X-Ray structure of human Aquaporin 2 crystallized on a silicon chip | | Descriptor: | Aquaporin-2, CADMIUM ION | | Authors: | Lieske, J, Cerv, M, Kreida, S, Barthelmess, M, Fischer, P, Pakendorf, T, Yefanov, O, Mariani, V, Seine, T, Ross, B.H, Crosas, E, Lorbeer, O, Burkhardt, A, Lane, T.J, Guenther, S, Bergtholdt, J, Schoen, S, Tornroth-Horsefield, S, Chapman, H.N, Meents, A. | | Deposit date: | 2019-01-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | On-chip crystallization for serial crystallography experiments and on-chip ligand-binding studies.
Iucrj, 6, 2019
|
|
7PVB
 
 | |
6D1Q
 
 | | Crystal structure of E. coli RppH-DapF complex, monomer | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, GLYCEROL, ... | | Authors: | Gao, A, Serganov, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and kinetic insights into stimulation of RppH-dependent RNA degradation by the metabolic enzyme DapF.
Nucleic Acids Res., 46, 2018
|
|
