8SG8
 
 | | CCT G beta 5 complex closed state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Guanine nucleotide-binding protein subunit beta-5, ... | | Authors: | Wang, S, Sass, M, Willardson, B.M, Shen, P.S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Visualizing the chaperone-mediated folding trajectory of the G protein beta 5 beta-propeller.
Mol.Cell, 83, 2023
|
|
8F4U
 
 | | Crystal structure of acetyltransferase Eis from M. tuberculosis in complex with azelastine | | Descriptor: | 4-[(4-chlorophenyl)methyl]-2-[(4S)-1-methylazepan-4-yl]phthalazin-1(2H)-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-11-11 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery and Mechanistic Analysis of Structurally Diverse Inhibitors of Acetyltransferase Eis among FDA-Approved Drugs.
Biochemistry, 62, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
9N1T
 
 | |
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | Descriptor: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8YKU
 
 | |
7SUS
 
 | | Crystal structure of Apelin receptor in complex with small molecule | | Descriptor: | (1R,2S)-N-[4-(2,6-dimethoxyphenyl)-5-(6-methylpyridin-2-yl)-1,2,4-triazol-3-yl]-1-(5-methylpyrimidin-2-yl)-1-oxidanyl-propane-2-sulfonamide, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apelin receptor, ... | | Authors: | Xu, F, Yue, Y, Liu, L.E, Han, G.W, Hanson, M. | | Deposit date: | 2021-11-18 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into apelin receptor-G protein stoichiometry.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SYP
 
 | | Structure of the wt IRES and 40S ribosome binary complex, open conformation. Structure 10(wt) | | Descriptor: | 18S rRNA, HCV IRES, HCV IRES partially loaded mRNA portion, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYQ
 
 | | Structure of the wt IRES and 40S ribosome ternary complex, open conformation. Structure 11(wt) | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYR
 
 | | Structure of the wt IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(wt). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8EPJ
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 17 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(3-{[4-(morpholin-4-yl)-2-nitroanilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
9NAI
 
 | | LuIII VLP - Glycan [s(Lex)2] | | Descriptor: | Capsid protein VP1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Busuttil, K.B, Bennett, A.B. | | Deposit date: | 2025-02-12 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Mapping the Sialic Acid Binding Sites of LuIII and H-1 Parvovirus
To Be Published
|
|
7SYS
 
 | | Structure of the delta dII IRES eIF2-containing 48S initiation complex, closed conformation. Structure 12(delta dII). | | Descriptor: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8EHP
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 13 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[(4-{[4-(morpholin-4-yl)anilino]methyl}phenyl)methyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EID
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 14 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({[(5P)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ELE
 
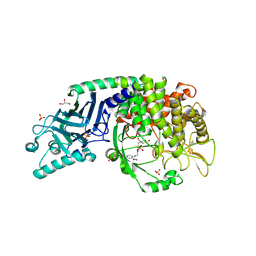 | | Co-crystal structure of Chaetomium glucosidase with compound 16 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[4-({2-nitro-4-[(1R,5S)-3-oxa-8-azabicyclo[3.2.1]octan-8-yl]anilino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-09-23 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ETL
 
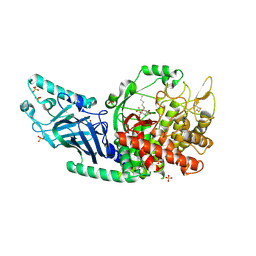 | | Co-crystal structure of Chaetomium glucosidase with compound 24 | | Descriptor: | (1S,2S,3R,4S,5S)-5-(butylamino)-1-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-17 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
5YDI
 
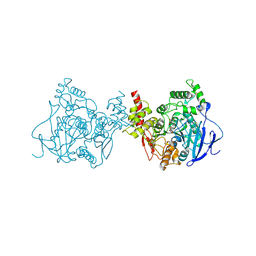 | | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector anopheles gambiae, new crystal packing | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Han, Q, Guan, H, Ding, H, Liao, C, Robinson, H, Li, J. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal structures of acetylcholinesterase of the malaria vector Anopheles gambiae reveal a polymerization interface, ligand binding residues and post translational modifications
To Be Published
|
|
8EUR
 
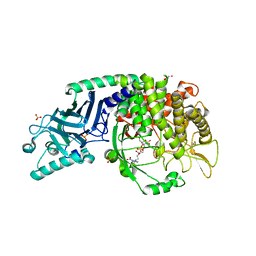 | | Co-crystal structure of Chaetomium glucosidase with compound 26 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{[2-nitro-4-(triazan-1-yl)phenyl]amino}ethyl (2-{[(1S,2S,3R,4S,5S)-2,3,4,5-tetrahydroxy-5-(hydroxymethyl)cyclohexyl]amino}ethyl)carbamate, Chaetomium alpha glucosidase, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EUT
 
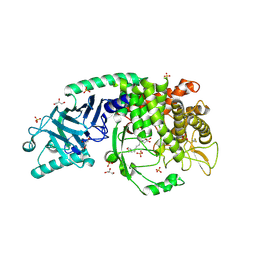 | | Co-crystal structure of Chaetomium glucosidase with compound 27 | | Descriptor: | (2R,3R,4R,5S)-1-[8-(furan-2-yl)octyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
9NAJ
 
 | | LuIII VLP - Glycan [3's(LN)3] | | Descriptor: | Capsid protein VP1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Busuttil, K.B, Bennett, A.B. | | Deposit date: | 2025-02-12 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Mapping the Sialic Acid Binding Sites of LuIII and H-1 Parvovirus
To Be Published
|
|
7Z5J
 
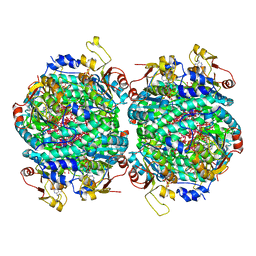 | | The molybdenum storage protein loaded with tungstate | | Descriptor: | 1,1,3,3,5,7,7,9,11,15,15-undecakis($l^{1}-oxidanyl)-2$l^{4},4$l^{3},6$l^{5},8,10,12,14,16,17,18,19$l^{3},20,21,22,23-pentadecaoxa-1$l^{6},3$l^{6},5$l^{6},7$l^{6},9$l^{6},11$l^{6},13$l^{6},15$l^{6}-octatungstapentadecacyclo[7.7.1.1^{1,13}.1^{3,5}.1^{3,15}.1^{5,7}.1^{5,11}.1^{7,11}.0^{2,13}.0^{2,15}.0^{4,13}.0^{6,9}.0^{6,11}.0^{6,13}.0^{9,19}]tricosane, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ermler, U, Aziz, I, Kaltwasser, S, Kayastha, K, Vonck, J. | | Deposit date: | 2022-03-09 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The molybdenum storage protein forms and deposits distinct polynuclear tungsten oxygen aggregates.
J.Inorg.Biochem., 234, 2022
|
|
8XVE
 
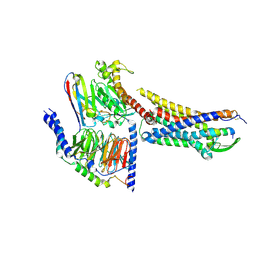 | | Cryo-EM structure of ETBR bound with BQ3020 | | Descriptor: | BQ3020, Exo-alpha-sialidase,Endothelin receptor type B, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hou, J.Y, Liu, S.H, Wu, L.J, Liu, Z.J, Hua, T. | | Deposit date: | 2024-01-15 | | Release date: | 2024-08-28 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of antagonist selectivity in endothelin receptors.
Cell Discov, 10, 2024
|
|
6ZG1
 
 | | SARM1 SAM1-2 domains | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sporny, M, Guez-Haddad, J, Khazma, T, Yaron, A, Dessau, M, Mim, C, Isupov, M.N, Zalk, R, Hons, M, Opatowsky, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2020-11-11 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for SARM1 inhibition and activation under energetic stress.
Elife, 9, 2020
|
|
