8ESQ
 
 | | Ytm1 associated nascent 60S ribosome State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUY
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETH
 
 | | Ytm1 associated 60S nascent ribosome State 1B | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ESR
 
 | | Ytm1 associated nascent 60S ribosome (-fkbp39) State 2 | | Descriptor: | 25S rRNA (cytosine-C(5))-methyltransferase nop2, 60S ribosomal protein L13, 60S ribosomal protein L14, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-14 | | Release date: | 2022-11-30 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8ETI
 
 | | Fkbp39 associated 60S nascent ribosome State 1 | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
8EUP
 
 | | Ytm1 associated 60S nascent ribosome State 1A | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L14, 60S ribosomal protein L15-A, ... | | Authors: | Zhou, X, Bilokapic, S, Deshmukh, A.A, Halic, M. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Chromatin localization of nucleophosmin organizes ribosome biogenesis.
Mol.Cell, 82, 2022
|
|
2XB2
 
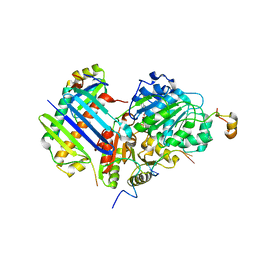 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JCM
 
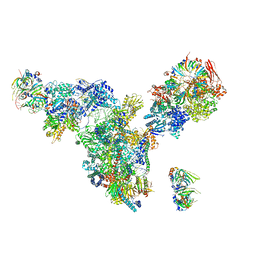 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
3JCR
 
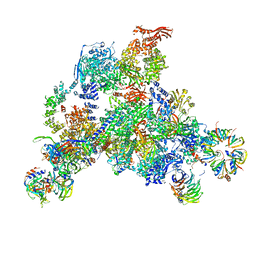 | | 3D structure determination of the human*U4/U6.U5* tri-snRNP complex | | Descriptor: | LSm2, LSm3, LSm4, ... | | Authors: | Agafonov, D.E, Kastner, B, Dybkov, O, Hofele, R.V, Liu, W.T, Urlaub, H, Luhrmann, R, Stark, H. | | Deposit date: | 2016-01-21 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular architecture of the human U4/U6.U5 tri-snRNP.
Science, 351, 2016
|
|
2J0S
 
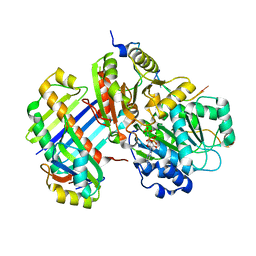 | | The crystal structure of the Exon Junction Complex at 2.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP *UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Mantains a Stable Grip on Mrna
Cell(Cambridge,Mass.), 126, 2006
|
|
2J0Q
 
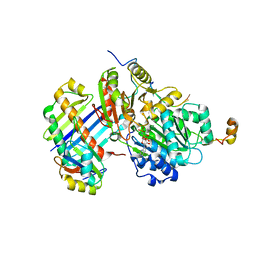 | | The crystal structure of the Exon Junction Complex at 3.2 A resolution | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-DEPENDENT RNA HELICASE DDX48, MAGNESIUM ION, ... | | Authors: | Bono, F, Ebert, J, Lorentzen, E, Conti, E. | | Deposit date: | 2006-08-04 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structure of the Exon Junction Complex Reveals How It Maintains a Stable Grip on Mrna.
Cell(Cambridge,Mass.), 126, 2006
|
|
2HYI
 
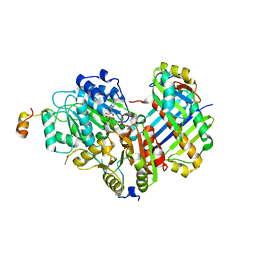 | | Structure of the human exon junction complex with a trapped DEAD-box helicase bound to RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*U)-3', MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andersen, C.B.F, Le Hir, H, Andersen, G.R. | | Deposit date: | 2006-08-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
7MQA
 
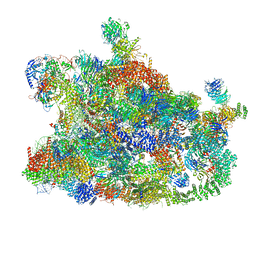 | | Cryo-EM structure of the human SSU processome, state post-A1 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Vanden Broeck, A, Singh, S, Klinge, S. | | Deposit date: | 2021-05-05 | | Release date: | 2021-09-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Nucleolar maturation of the human small subunit processome.
Science, 373, 2021
|
|
6ICZ
 
 | | Cryo-EM structure of a human post-catalytic spliceosome (P complex) at 3.0 angstrom | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
3EX7
 
 | |
7NAC
 
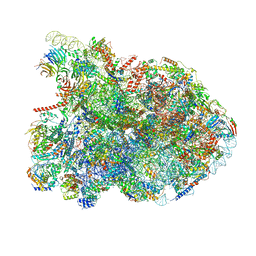 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Composite model | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NAD
 
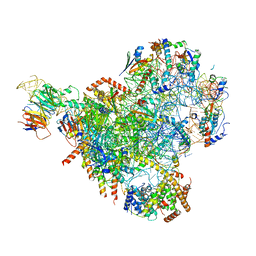 | | State E2 nucleolar 60S ribosomal biogenesis intermediate - Spb4 local refinement model | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L17-A, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2021-06-21 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OI6
 
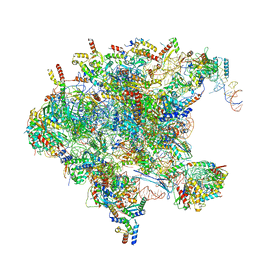 | | Cryo-EM structure of late human 39S mitoribosome assembly intermediates, state 1 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-13 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A distinct assembly pathway of the human 39S late pre-mitoribosome.
Nat Commun, 12, 2021
|
|
7OHS
 
 | |
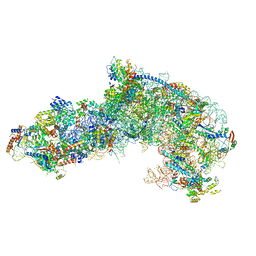
7OHW
 
 | |
7OHP
 
 | |
7OHV
 
 | |
7OHX
 
 | |
7OHR
 
 | | Nog1-TAP associated immature ribosomal particle population E from S. cerevisiae | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.72 Å) | | Cite: | Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors.
Plos One, 16, 2021
|
|
6ID1
 
 | | Cryo-EM structure of a human intron lariat spliceosome after Prp43 loaded (ILS2 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
